1.1. Calculating landcover distribution & vegetation extraction
Farzad Vahidi Mayamey
Aim of the studies
Calculating landcover distribution in our wetlands based on 10 m landcover map of Sweden
Open water and flooded vegetation extraction in one of our wetlands.
1.1.1. Calculating landcover distribution
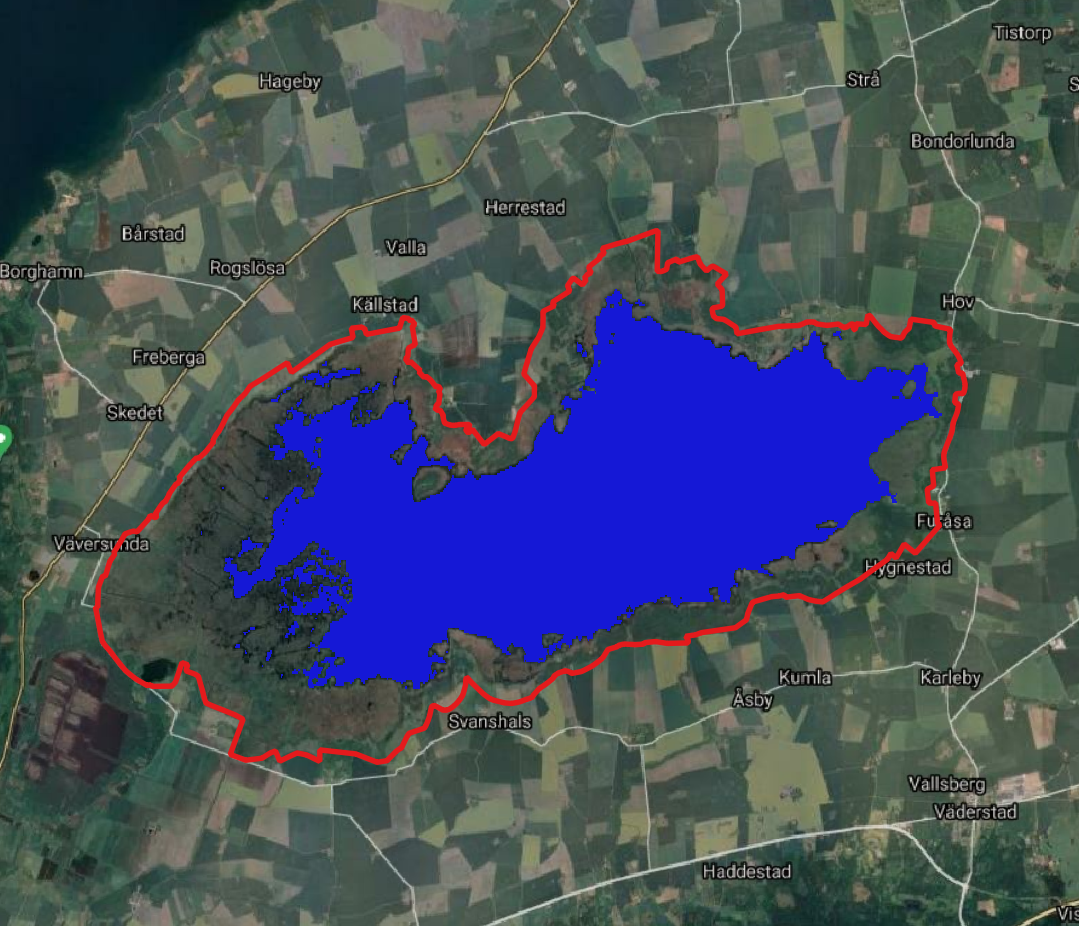
we use two different layer: 1. National landcover map of Sweden in 10m resolution ( left picture) 2. vector layer of Ramsar sites in Sweden ( right picture)
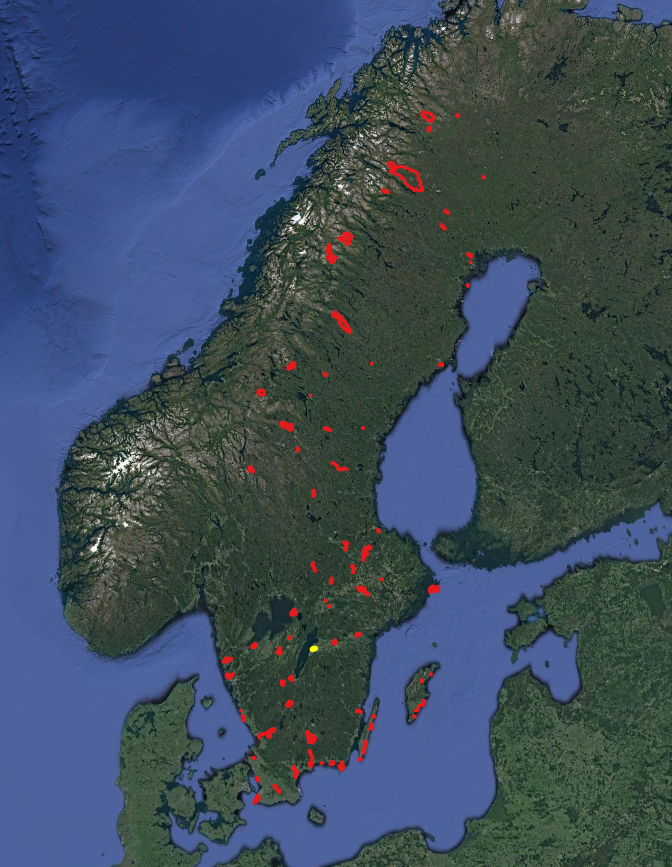

[100]:
%%bash
gdalinfo nmd2018bas_ogeneraliserad_v1_1.tif | more
Driver: GTiff/GeoTIFF
Files: nmd2018bas_ogeneraliserad_v1_1.tif
Size is 71273, 157992
Coordinate System is:
PROJCS["SWEREF99 TM",
GEOGCS["SWEREF99",
DATUM["SWEREF99",
SPHEROID["GRS 1980",6378137,298.257222101,
AUTHORITY["EPSG","7019"]],
TOWGS84[0,0,0,0,0,0,0],
AUTHORITY["EPSG","6619"]],
PRIMEM["Greenwich",0,
AUTHORITY["EPSG","8901"]],
UNIT["degree",0.0174532925199433,
AUTHORITY["EPSG","9122"]],
AUTHORITY["EPSG","4619"]],
PROJECTION["Transverse_Mercator"],
PARAMETER["latitude_of_origin",0],
PARAMETER["central_meridian",15],
PARAMETER["scale_factor",0.9996],
PARAMETER["false_easting",500000],
PARAMETER["false_northing",0],
UNIT["metre",1,
AUTHORITY["EPSG","9001"]],
AUTHORITY["EPSG","3006"]]
Origin = (208450.000000000000000,7671060.000000000000000)
Pixel Size = (10.000000000000000,-10.000000000000000)
Metadata:
AREA_OR_POINT=Area
TIFFTAG_RESOLUTIONUNIT=2 (pixels/inch)
TIFFTAG_SOFTWARE=E
TIFFTAG_XRESOLUTION=1
TIFFTAG_YRESOLUTION=1
Image Structure Metadata:
COMPRESSION=PACKBITS
INTERLEAVE=BAND
Corner Coordinates:
Upper Left ( 208450.000, 7671060.000) ( 7d41'51.00"E, 68d59'38.16"N)
Lower Left ( 208450.000, 6091140.000) ( 10d27'14.37"E, 54d52'55.53"N)
Upper Right ( 921180.000, 7671060.000) ( 25d29'31.13"E, 68d49'34.01"N)
Lower Right ( 921180.000, 6091140.000) ( 21d33'17.33"E, 54d47'23.89"N)
Center ( 564815.000, 6881100.000) ( 16d14'23.34"E, 62d 3'23.60"N)
Band 1 Block=512x512 Type=Byte, ColorInterp=Palette
NoData Value=0
Color Table (RGB with 256 entries)
0: 0,0,0,0
1: 0,0,0,255
2: 194,158,215,255
3: 255,255,190,255
4: 0,0,0,255
5: 0,0,0,255
6: 0,0,0,255
7: 0,0,0,255
8: 0,0,0,255
9: 0,0,0,255
10: 0,0,0,255
11: 0,0,0,255
12: 0,0,0,255
13: 0,0,0,255
14: 0,0,0,255
15: 0,0,0,255
16: 0,0,0,255
17: 0,0,0,255
18: 0,0,0,255
19: 0,0,0,255
20: 0,0,0,255
21: 0,0,0,255
22: 0,0,0,255
23: 0,0,0,255
24: 0,0,0,255
25: 0,0,0,255
26: 0,0,0,255
27: 0,0,0,255
28: 0,0,0,255
29: 0,0,0,255
30: 0,0,0,255
31: 0,0,0,255
32: 0,0,0,255
33: 0,0,0,255
34: 0,0,0,255
35: 0,0,0,255
36: 0,0,0,255
37: 0,0,0,255
38: 0,0,0,255
39: 0,0,0,255
40: 0,0,0,255
41: 225,225,225,255
42: 255,211,127,255
43: 0,0,0,255
44: 0,0,0,255
45: 0,0,0,255
46: 0,0,0,255
47: 0,0,0,255
48: 0,0,0,255
49: 0,0,0,255
50: 0,0,0,255
51: 90,20,20,255
52: 229,70,75,255
53: 25,25,25,255
54: 0,0,0,255
55: 0,0,0,255
56: 0,0,0,255
57: 0,0,0,255
58: 0,0,0,255
59: 0,0,0,255
60: 0,0,0,255
61: 102,153,205,255
62: 138,204,250,255
63: 0,0,0,255
64: 0,0,0,255
65: 0,0,0,255
66: 0,0,0,255
67: 0,0,0,255
68: 0,0,0,255
69: 0,0,0,255
70: 0,0,0,255
71: 0,0,0,255
72: 0,0,0,255
73: 0,0,0,255
74: 0,0,0,255
75: 0,0,0,255
76: 0,0,0,255
77: 0,0,0,255
78: 0,0,0,255
79: 0,0,0,255
80: 0,0,0,255
81: 0,0,0,255
82: 0,0,0,255
83: 0,0,0,255
84: 0,0,0,255
85: 0,0,0,255
86: 0,0,0,255
87: 0,0,0,255
88: 0,0,0,255
89: 0,0,0,255
90: 0,0,0,255
91: 0,0,0,255
92: 0,0,0,255
93: 0,0,0,255
94: 0,0,0,255
95: 0,0,0,255
96: 0,0,0,255
97: 0,0,0,255
98: 0,0,0,255
99: 0,0,0,255
100: 0,0,0,255
101: 0,0,0,255
102: 0,0,0,255
103: 0,0,0,255
104: 0,0,0,255
105: 0,0,0,255
106: 0,0,0,255
107: 0,0,0,255
108: 0,0,0,255
109: 0,0,0,255
110: 0,0,0,255
111: 110,140,5,255
112: 45,95,0,255
113: 78,112,0,255
114: 56,168,0,255
115: 76,230,0,255
116: 170,255,0,255
117: 151,230,0,255
118: 205,205,102,255
119: 0,0,0,255
120: 0,0,0,255
121: 89,140,85,255
122: 48,94,80,255
123: 35,115,90,255
124: 67,136,112,255
125: 137,205,155,255
126: 165,245,120,255
127: 171,205,120,255
128: 137,137,68,255
129: 0,0,0,255
130: 194,158,215,255
131: 255,255,190,255
132: 0,0,0,255
133: 0,0,0,255
134: 0,0,0,255
135: 0,0,0,255
136: 0,0,0,255
137: 0,0,0,255
138: 0,0,0,255
139: 0,0,0,255
140: 0,0,0,255
141: 0,0,0,255
142: 0,0,0,255
143: 0,0,0,255
144: 0,0,0,255
145: 0,0,0,255
146: 0,0,0,255
147: 0,0,0,255
148: 0,0,0,255
149: 0,0,0,255
150: 0,0,0,255
151: 0,0,0,255
152: 0,0,0,255
153: 0,0,0,255
154: 0,0,0,255
155: 0,0,0,255
156: 0,0,0,255
157: 0,0,0,255
158: 0,0,0,255
159: 0,0,0,255
160: 0,0,0,255
161: 0,0,0,255
162: 0,0,0,255
163: 0,0,0,255
164: 0,0,0,255
165: 0,0,0,255
166: 0,0,0,255
167: 0,0,0,255
168: 0,0,0,255
169: 225,225,225,255
170: 255,211,127,255
171: 0,0,0,255
172: 0,0,0,255
173: 0,0,0,255
174: 0,0,0,255
175: 0,0,0,255
176: 0,0,0,255
177: 0,0,0,255
178: 0,0,0,255
179: 90,20,20,255
180: 229,70,75,255
181: 25,25,25,255
182: 0,0,0,255
183: 0,0,0,255
184: 0,0,0,255
185: 0,0,0,255
186: 0,0,0,255
187: 0,0,0,255
188: 0,0,0,255
189: 102,153,205,255
190: 138,204,250,255
191: 0,0,0,255
192: 192,192,192,255
193: 193,193,193,255
194: 194,194,194,255
195: 195,195,195,255
196: 196,196,196,255
197: 197,197,197,255
198: 198,198,198,255
199: 199,199,199,255
200: 200,200,200,255
201: 201,201,201,255
202: 202,202,202,255
203: 203,203,203,255
204: 204,204,204,255
205: 205,205,205,255
206: 206,206,206,255
207: 207,207,207,255
208: 208,208,208,255
209: 209,209,209,255
210: 210,210,210,255
211: 211,211,211,255
212: 212,212,212,255
213: 213,213,213,255
214: 214,214,214,255
215: 215,215,215,255
216: 216,216,216,255
217: 217,217,217,255
218: 218,218,218,255
219: 219,219,219,255
220: 220,220,220,255
221: 221,221,221,255
222: 222,222,222,255
223: 223,223,223,255
224: 224,224,224,255
225: 225,225,225,255
226: 226,226,226,255
227: 227,227,227,255
228: 228,228,228,255
229: 229,229,229,255
230: 230,230,230,255
231: 231,231,231,255
232: 232,232,232,255
233: 233,233,233,255
234: 234,234,234,255
235: 235,235,235,255
236: 236,236,236,255
237: 237,237,237,255
238: 238,238,238,255
239: 239,239,239,255
240: 240,240,240,255
241: 241,241,241,255
242: 242,242,242,255
243: 243,243,243,255
244: 244,244,244,255
245: 245,245,245,255
246: 246,246,246,255
247: 247,247,247,255
248: 248,248,248,255
249: 249,249,249,255
250: 250,250,250,255
251: 251,251,251,255
252: 252,252,252,255
253: 253,253,253,255
254: 254,254,254,255
255: 255,255,255,255
As NoData Value is not assigned, we assign nodata value = 0
[102]:
%%bash
gdal_edit.py -a_nodata 0 nmd2018bas_ogeneraliserad_v1_1.tif
we create the shapefile of each wetland by using $ split vector layer tool in QGIS to clip our landcover by this layers
[104]:
%%bash
for file in rms/*.gpkg; do echo $file; filename=$(basename $file .gpkg); gdalwarp -overwrite -srcnodata 0 -dstnodata 0 -crop_to_cutline -cutline $file nmd2018bas_ogeneraliserad_v1_1.tif clipped/${filename}.tif; done
rms/ramsarid_1113.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 2623P x 1832L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1114.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 2745P x 1850L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1115.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 4953P x 1980L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1116.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 2032P x 3435L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1117.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 694P x 1137L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1118.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 997P x 633L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1119.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1370P x 1214L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1120.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 421P x 661L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1121.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 807P x 1246L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1122.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 636P x 1020L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1123.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 835P x 3896L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1124.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 497P x 421L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1125.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1724P x 1163L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1126.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1579P x 1571L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1127.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 725P x 371L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1128.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1269P x 691L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1129.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1074P x 1192L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1130.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 376P x 444L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1131.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1803P x 2087L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1132.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 265P x 323L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_1133.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 857P x 1257L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_14.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1215P x 2045L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_15.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1538P x 1692L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_16.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1224P x 2753L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_17.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 567P x 663L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_18.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 2657P x 8279L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_19.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 299P x 262L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_20.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1340P x 1415L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2167.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 4054P x 4742L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2168.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 544P x 1222L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2169.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 359P x 1003L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2170.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 347P x 776L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2171.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 546P x 1439L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2172.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 364P x 521L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2173.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 3518P x 1881L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2174.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 253P x 289L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2175.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1224P x 2245L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2176.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1229P x 890L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2177.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 739P x 1017L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2178.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1124P x 1273L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2179.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 366P x 262L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2180.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1658P x 795L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_21.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 3979P x 9467L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2260.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 838P x 618L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_2265.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 236P x 441L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_22.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1237P x 1576L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_23.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1250P x 763L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_24.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 585P x 386L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_25.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 310P x 437L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_26.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1753P x 1250L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_27.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 355P x 376L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_28.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1136P x 1172L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_29.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1933P x 4352L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_30.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 3144P x 2662L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_31.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1719P x 712L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_32.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 7512P x 6180L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_33.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 2602P x 2191L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_429.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1831P x 2819L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_430.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 647P x 1604L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_431.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1962P x 1036L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_432.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1120P x 1022L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_433.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 539P x 694L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_434.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1248P x 1514L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_435.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 2131P x 1433L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_436.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 937P x 1652L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_437.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 1353P x 2170L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
rms/ramsarid_438.gpkg
Copying color table from nmd2018bas_ogeneraliserad_v1_1.tif to new file.
Creating output file that is 687P x 666L.
Processing nmd2018bas_ogeneraliserad_v1_1.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
Now that we have our clipped land cover we can calculate the histogram (in percentage)
[105]:
%%bash
for file in clipped/*.tif; do echo $file; filename=$(basename $file .tif); pkstat -hist -rel -src_min 2 -src_max 128 -i $file > hist/${filename}.txt; done
clipped/ramsarid_1113.tif
clipped/ramsarid_1114.tif
clipped/ramsarid_1115.tif
clipped/ramsarid_1116.tif
clipped/ramsarid_1117.tif
clipped/ramsarid_1118.tif
clipped/ramsarid_1119.tif
clipped/ramsarid_1120.tif
clipped/ramsarid_1121.tif
clipped/ramsarid_1122.tif
clipped/ramsarid_1123.tif
clipped/ramsarid_1124.tif
clipped/ramsarid_1125.tif
clipped/ramsarid_1126.tif
clipped/ramsarid_1127.tif
clipped/ramsarid_1128.tif
clipped/ramsarid_1129.tif
clipped/ramsarid_1130.tif
clipped/ramsarid_1131.tif
clipped/ramsarid_1132.tif
clipped/ramsarid_1133.tif
clipped/ramsarid_14.tif
clipped/ramsarid_15.tif
clipped/ramsarid_16.tif
clipped/ramsarid_17.tif
clipped/ramsarid_18.tif
clipped/ramsarid_19.tif
clipped/ramsarid_20.tif
clipped/ramsarid_2167.tif
clipped/ramsarid_2168.tif
clipped/ramsarid_2169.tif
clipped/ramsarid_2170.tif
clipped/ramsarid_2171.tif
clipped/ramsarid_2172.tif
clipped/ramsarid_2173.tif
clipped/ramsarid_2174.tif
clipped/ramsarid_2175.tif
clipped/ramsarid_2176.tif
clipped/ramsarid_2177.tif
clipped/ramsarid_2178.tif
clipped/ramsarid_2179.tif
clipped/ramsarid_2180.tif
clipped/ramsarid_21.tif
clipped/ramsarid_2260.tif
clipped/ramsarid_2265.tif
clipped/ramsarid_22.tif
clipped/ramsarid_23.tif
clipped/ramsarid_24.tif
clipped/ramsarid_25.tif
clipped/ramsarid_26.tif
clipped/ramsarid_27.tif
clipped/ramsarid_28.tif
clipped/ramsarid_29.tif
clipped/ramsarid_30.tif
clipped/ramsarid_31.tif
clipped/ramsarid_32.tif
clipped/ramsarid_33.tif
clipped/ramsarid_429.tif
clipped/ramsarid_430.tif
clipped/ramsarid_431.tif
clipped/ramsarid_432.tif
clipped/ramsarid_433.tif
clipped/ramsarid_434.tif
clipped/ramsarid_435.tif
clipped/ramsarid_436.tif
clipped/ramsarid_437.tif
clipped/ramsarid_438.tif
Now we have the text file of our histograms we can extract our desired classes for each of our 67 wetlands - we have 25 classess in total - 7 of the classess are our main interest
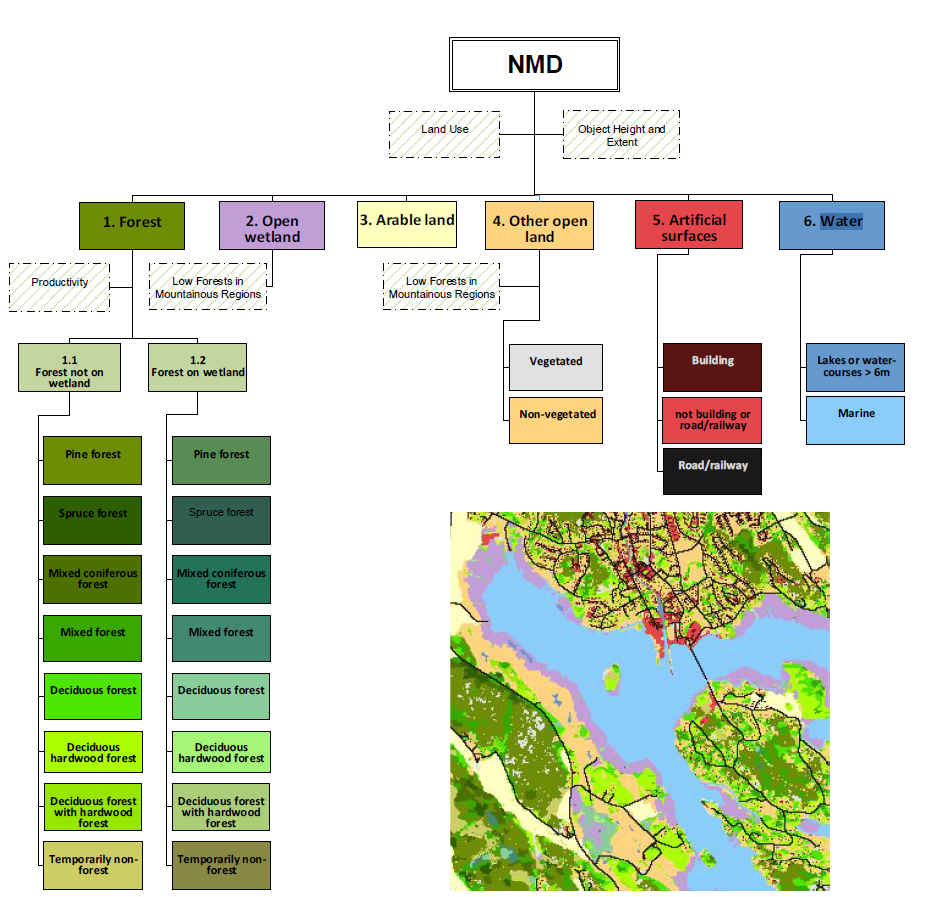
Extract all 25 classes
[106]:
%%bash
#printf "%s %.3f\n", $1, $2
for file in hist/*.txt; do echo $file; filename=$(basename $file .txt);
awk '{ if($1==111||$1==112||$1==113||$1==114||$1==115||$1==116||$1==117||$1==118||$1==121||$1==122||$1==123||$1==124||$1==125||$1==126||$1==127||$1==128||$1==2||$1==3||$1==41||$1==42||$1==51||$1==52||$1==53||$1==61||$1==62) print}' $file > class_25/${filename}.txt;done
hist/ramsarid_1113.txt
hist/ramsarid_1114.txt
hist/ramsarid_1115.txt
hist/ramsarid_1116.txt
hist/ramsarid_1117.txt
hist/ramsarid_1118.txt
hist/ramsarid_1119.txt
hist/ramsarid_1120.txt
hist/ramsarid_1121.txt
hist/ramsarid_1122.txt
hist/ramsarid_1123.txt
hist/ramsarid_1124.txt
hist/ramsarid_1125.txt
hist/ramsarid_1126.txt
hist/ramsarid_1127.txt
hist/ramsarid_1128.txt
hist/ramsarid_1129.txt
hist/ramsarid_1130.txt
hist/ramsarid_1131.txt
hist/ramsarid_1132.txt
hist/ramsarid_1133.txt
hist/ramsarid_14.txt
hist/ramsarid_15.txt
hist/ramsarid_16.txt
hist/ramsarid_17.txt
hist/ramsarid_18.txt
hist/ramsarid_19.txt
hist/ramsarid_20.txt
hist/ramsarid_2167.txt
hist/ramsarid_2168.txt
hist/ramsarid_2169.txt
hist/ramsarid_2170.txt
hist/ramsarid_2171.txt
hist/ramsarid_2172.txt
hist/ramsarid_2173.txt
hist/ramsarid_2174.txt
hist/ramsarid_2175.txt
hist/ramsarid_2176.txt
hist/ramsarid_2177.txt
hist/ramsarid_2178.txt
hist/ramsarid_2179.txt
hist/ramsarid_2180.txt
hist/ramsarid_21.txt
hist/ramsarid_2260.txt
hist/ramsarid_2265.txt
hist/ramsarid_22.txt
hist/ramsarid_23.txt
hist/ramsarid_24.txt
hist/ramsarid_25.txt
hist/ramsarid_26.txt
hist/ramsarid_27.txt
hist/ramsarid_28.txt
hist/ramsarid_29.txt
hist/ramsarid_30.txt
hist/ramsarid_31.txt
hist/ramsarid_32.txt
hist/ramsarid_33.txt
hist/ramsarid_429.txt
hist/ramsarid_430.txt
hist/ramsarid_431.txt
hist/ramsarid_432.txt
hist/ramsarid_433.txt
hist/ramsarid_434.txt
hist/ramsarid_435.txt
hist/ramsarid_436.txt
hist/ramsarid_437.txt
hist/ramsarid_438.txt
Extract 7 main classess of our interest
[107]:
%%bash
for file in hist/*.txt; do echo $file; filename=$(basename $file .txt);
awk '$1==111 || $1==112 || $1==113 || $1==114 ||$1==115 || $1==116 ||$1==117 || $1==118 {sum1+= $2}
$1==121 || $1==122 || $1==123 || $1==124 ||$1==125 || $1==126 ||$1==127 || $1==128 {sum2+= $2}
$1==2 {sum3+= $2}
$1==3 {sum4+= $2}
$1==41 || $1==42 {sum5+= $2}
$1==51 || $1==52 || $1==53 {sum6+= $2}
$1==61 || $1==62 {sum7+= $2}
END{print ("1.1 "sum1); print ("1.2 "sum2); print("2 "sum3); print("3 "sum4); print("4 "sum5); print("5 "sum6); print("6 "sum7)}' $file > class_7/${filename}.txt;done
hist/ramsarid_1113.txt
hist/ramsarid_1114.txt
hist/ramsarid_1115.txt
hist/ramsarid_1116.txt
hist/ramsarid_1117.txt
hist/ramsarid_1118.txt
hist/ramsarid_1119.txt
hist/ramsarid_1120.txt
hist/ramsarid_1121.txt
hist/ramsarid_1122.txt
hist/ramsarid_1123.txt
hist/ramsarid_1124.txt
hist/ramsarid_1125.txt
hist/ramsarid_1126.txt
hist/ramsarid_1127.txt
hist/ramsarid_1128.txt
hist/ramsarid_1129.txt
hist/ramsarid_1130.txt
hist/ramsarid_1131.txt
hist/ramsarid_1132.txt
hist/ramsarid_1133.txt
hist/ramsarid_14.txt
hist/ramsarid_15.txt
hist/ramsarid_16.txt
hist/ramsarid_17.txt
hist/ramsarid_18.txt
hist/ramsarid_19.txt
hist/ramsarid_20.txt
hist/ramsarid_2167.txt
hist/ramsarid_2168.txt
hist/ramsarid_2169.txt
hist/ramsarid_2170.txt
hist/ramsarid_2171.txt
hist/ramsarid_2172.txt
hist/ramsarid_2173.txt
hist/ramsarid_2174.txt
hist/ramsarid_2175.txt
hist/ramsarid_2176.txt
hist/ramsarid_2177.txt
hist/ramsarid_2178.txt
hist/ramsarid_2179.txt
hist/ramsarid_2180.txt
hist/ramsarid_21.txt
hist/ramsarid_2260.txt
hist/ramsarid_2265.txt
hist/ramsarid_22.txt
hist/ramsarid_23.txt
hist/ramsarid_24.txt
hist/ramsarid_25.txt
hist/ramsarid_26.txt
hist/ramsarid_27.txt
hist/ramsarid_28.txt
hist/ramsarid_29.txt
hist/ramsarid_30.txt
hist/ramsarid_31.txt
hist/ramsarid_32.txt
hist/ramsarid_33.txt
hist/ramsarid_429.txt
hist/ramsarid_430.txt
hist/ramsarid_431.txt
hist/ramsarid_432.txt
hist/ramsarid_433.txt
hist/ramsarid_434.txt
hist/ramsarid_435.txt
hist/ramsarid_436.txt
hist/ramsarid_437.txt
hist/ramsarid_438.txt
[108]:
%%bash
for file in class_7/*.txt; do echo $file; filename=$(basename $file .txt);
awk 'BEGIN{print "Class Ratio"}1' $file > head/${filename}.txt; done #The 1 is to indicate to print every line of the file.
class_7/ramsarid_1113.txt
class_7/ramsarid_1114.txt
class_7/ramsarid_1115.txt
class_7/ramsarid_1116.txt
class_7/ramsarid_1117.txt
class_7/ramsarid_1118.txt
class_7/ramsarid_1119.txt
class_7/ramsarid_1120.txt
class_7/ramsarid_1121.txt
class_7/ramsarid_1122.txt
class_7/ramsarid_1123.txt
class_7/ramsarid_1124.txt
class_7/ramsarid_1125.txt
class_7/ramsarid_1126.txt
class_7/ramsarid_1127.txt
class_7/ramsarid_1128.txt
class_7/ramsarid_1129.txt
class_7/ramsarid_1130.txt
class_7/ramsarid_1131.txt
class_7/ramsarid_1132.txt
class_7/ramsarid_1133.txt
class_7/ramsarid_14.txt
class_7/ramsarid_15.txt
class_7/ramsarid_16.txt
class_7/ramsarid_17.txt
class_7/ramsarid_18.txt
class_7/ramsarid_19.txt
class_7/ramsarid_20.txt
class_7/ramsarid_2167.txt
class_7/ramsarid_2168.txt
class_7/ramsarid_2169.txt
class_7/ramsarid_2170.txt
class_7/ramsarid_2171.txt
class_7/ramsarid_2172.txt
class_7/ramsarid_2173.txt
class_7/ramsarid_2174.txt
class_7/ramsarid_2175.txt
class_7/ramsarid_2176.txt
class_7/ramsarid_2177.txt
class_7/ramsarid_2178.txt
class_7/ramsarid_2179.txt
class_7/ramsarid_2180.txt
class_7/ramsarid_21.txt
class_7/ramsarid_2260.txt
class_7/ramsarid_2265.txt
class_7/ramsarid_22.txt
class_7/ramsarid_23.txt
class_7/ramsarid_24.txt
class_7/ramsarid_25.txt
class_7/ramsarid_26.txt
class_7/ramsarid_27.txt
class_7/ramsarid_28.txt
class_7/ramsarid_29.txt
class_7/ramsarid_30.txt
class_7/ramsarid_31.txt
class_7/ramsarid_32.txt
class_7/ramsarid_33.txt
class_7/ramsarid_429.txt
class_7/ramsarid_430.txt
class_7/ramsarid_431.txt
class_7/ramsarid_432.txt
class_7/ramsarid_433.txt
class_7/ramsarid_434.txt
class_7/ramsarid_435.txt
class_7/ramsarid_436.txt
class_7/ramsarid_437.txt
class_7/ramsarid_438.txt
[76]:
pip install matplotlib
Collecting matplotlib
Downloading matplotlib-3.4.2-cp38-cp38-manylinux1_x86_64.whl (10.3 MB)
|████████████████████████████████| 10.3 MB 7.9 MB/s eta 0:00:01
Collecting pillow>=6.2.0
Downloading Pillow-8.2.0-cp38-cp38-manylinux1_x86_64.whl (3.0 MB)
|████████████████████████████████| 3.0 MB 17.5 MB/s eta 0:00:01
Requirement already satisfied: numpy>=1.16 in /home/user/miniconda3/lib/python3.8/site-packages (from matplotlib) (1.20.1)
Collecting cycler>=0.10
Downloading cycler-0.10.0-py2.py3-none-any.whl (6.5 kB)
Requirement already satisfied: python-dateutil>=2.7 in /home/user/miniconda3/lib/python3.8/site-packages (from matplotlib) (2.8.1)
Collecting kiwisolver>=1.0.1
Downloading kiwisolver-1.3.1-cp38-cp38-manylinux1_x86_64.whl (1.2 MB)
|████████████████████████████████| 1.2 MB 54.3 MB/s eta 0:00:01
Requirement already satisfied: pyparsing>=2.2.1 in /home/user/miniconda3/lib/python3.8/site-packages (from matplotlib) (2.4.7)
Requirement already satisfied: six in /home/user/miniconda3/lib/python3.8/site-packages (from cycler>=0.10->matplotlib) (1.15.0)
Installing collected packages: pillow, cycler, kiwisolver, matplotlib
Successfully installed cycler-0.10.0 kiwisolver-1.3.1 matplotlib-3.4.2 pillow-8.2.0
Note: you may need to restart the kernel to use updated packages.
[110]:
import matplotlib.pyplot as plt
import os
import glob
file_location = os.path.join('class_7', '*.txt')
filenames = glob.glob(file_location)
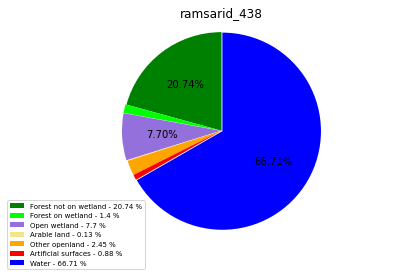
class_ = ['Forest not on wetland', 'Forest on wetland',
'Open wetland','Arable land', 'Other openland', 'Artificial surfaces',
'Water']
for f in filenames:
ratio = []
outfile = open(f,'r')
for line in outfile:
Data = [i for i in line.split()]
New_Data= [ j for j in Data[1].split()]
ratio.append(New_Data[0])
outfile.close()
file_name = os.path.basename(f)
file_name=os.path.splitext(file_name)[0]
print(file_name)
ratio = [float(i) for i in ratio]
#print(ratio)
colors = ['green', 'lime', 'mediumpurple', 'khaki','orange', 'red', 'blue']
explode = [0, 0.01, 0.01, 0, 0, 0, 0.01]
# plotting pie chart
def my_autopct(pct):
return ('%.2f%%' % pct) if pct > 5 else ''
plt.pie(ratio, colors = colors, startangle = 90, explode = explode,
shadow = False, autopct=my_autopct )
plt.title(label=file_name, loc='center')
ratio_r = [round(num, 2) for num in ratio]
labels = ['{0} - {1} %'.format(i, j) for i, j in zip(class_,ratio_r)]
plt.legend( labels, loc='lower left', bbox_to_anchor=(-0.15, -.15), fontsize=7)
plt.axis('equal')
#plt.tight_layout()
file_name_ext=os.path.splitext(file_name)[0]+".png"
plt.savefig(file_name_ext, format="png", dpi=300)
plt.show()
ramsarid_1113
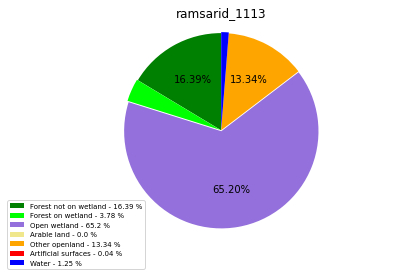
ramsarid_1114
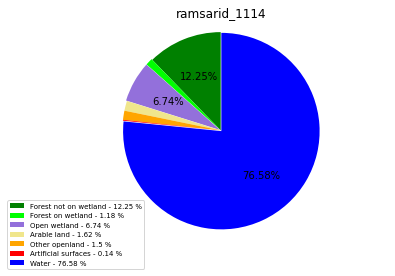
ramsarid_1115
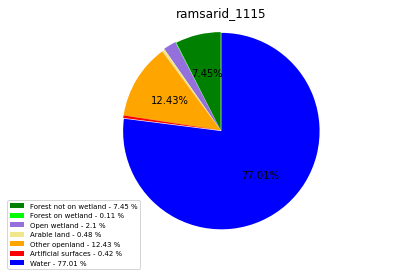
ramsarid_1116
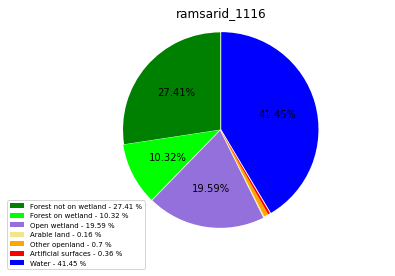
ramsarid_1117
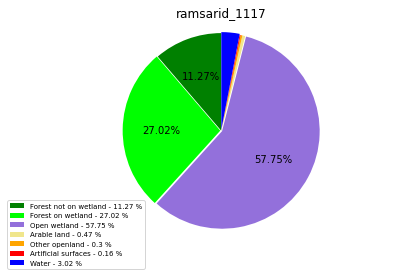
ramsarid_1118
ramsarid_1119
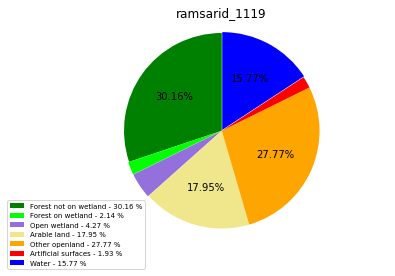
ramsarid_1120
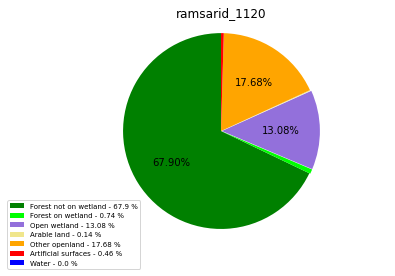
ramsarid_1121
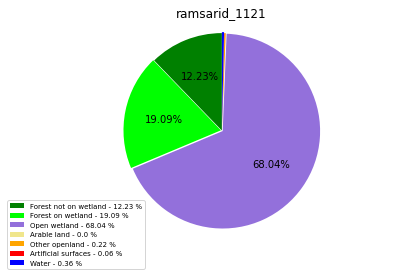
ramsarid_1122
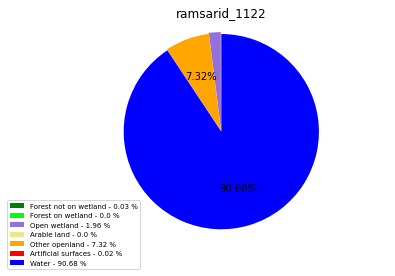
ramsarid_1123
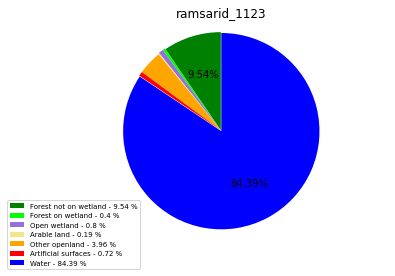
ramsarid_1124
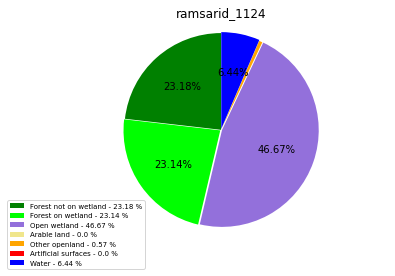
ramsarid_1125
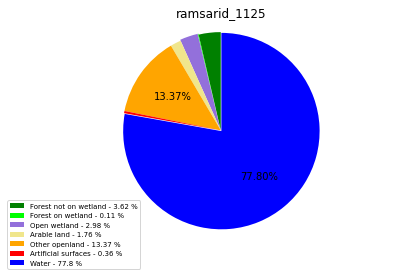
ramsarid_1126
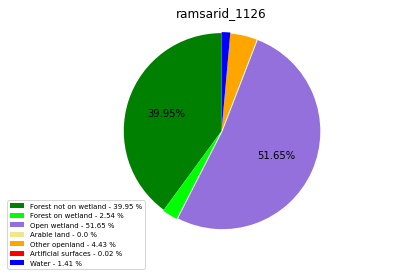
ramsarid_1127
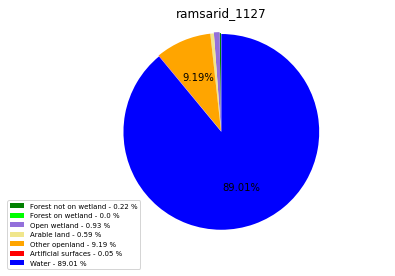
ramsarid_1128
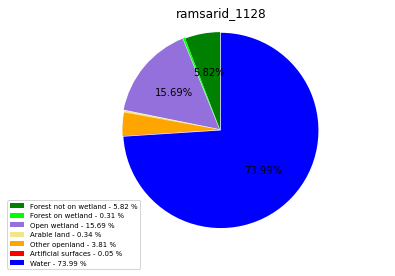
ramsarid_1129
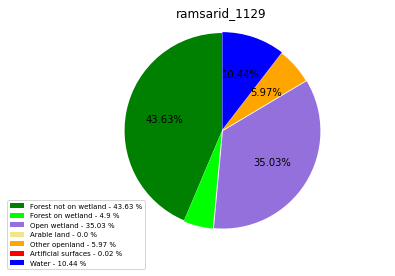
ramsarid_1130
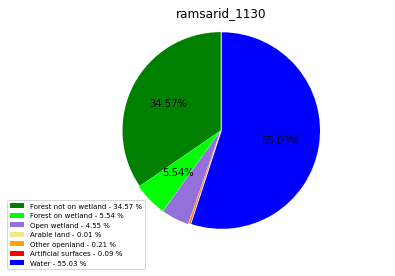
ramsarid_1131
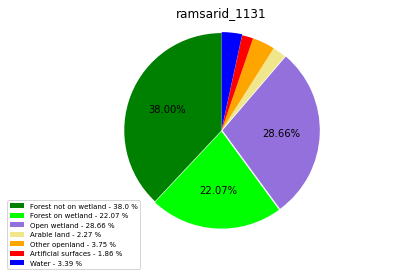
ramsarid_1132
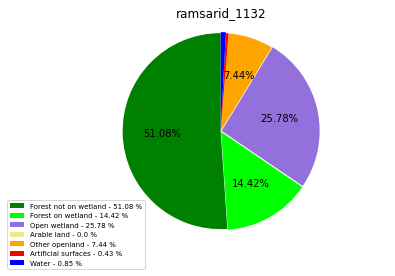
ramsarid_1133
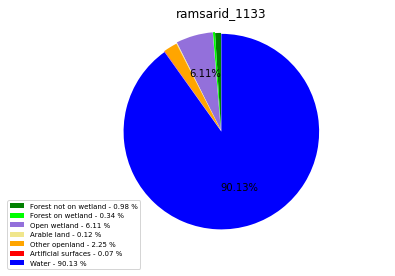
ramsarid_14
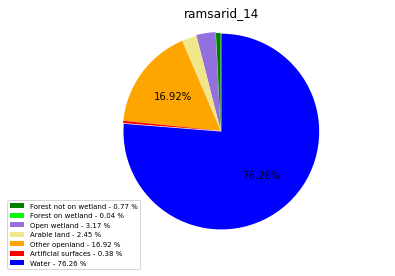
ramsarid_15
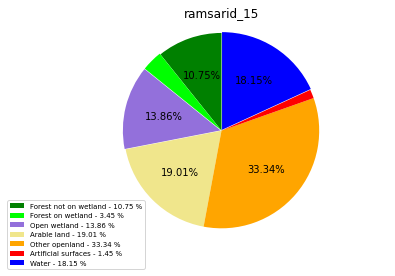
ramsarid_16
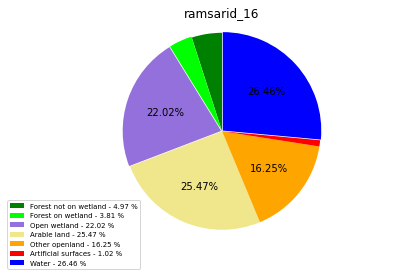
ramsarid_17
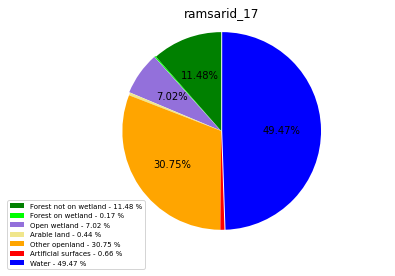
ramsarid_18
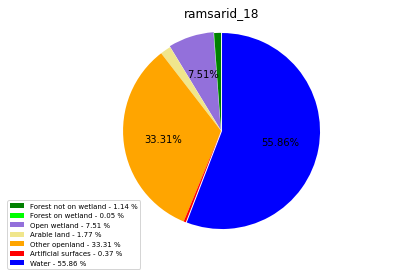
ramsarid_19
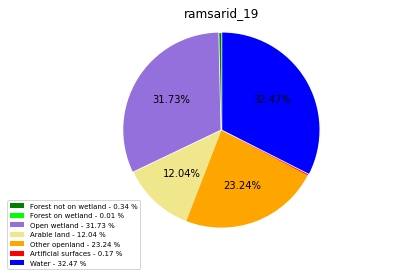
ramsarid_20
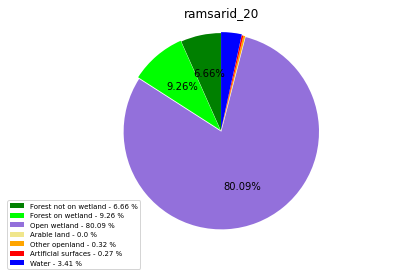
ramsarid_21
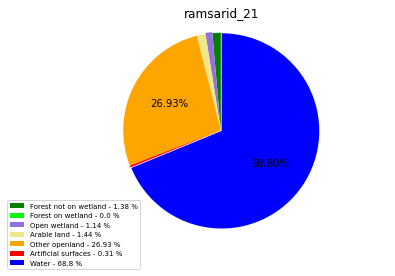
ramsarid_2167
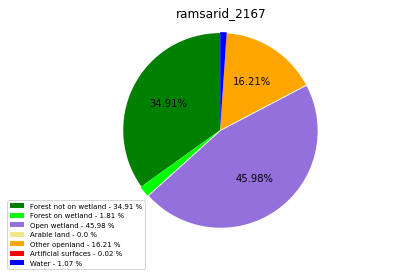
ramsarid_2168
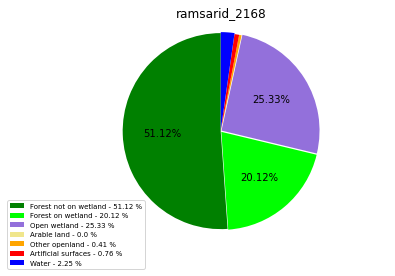
ramsarid_2169
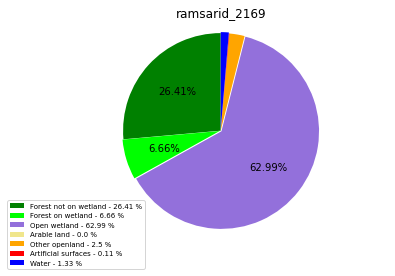
ramsarid_2170
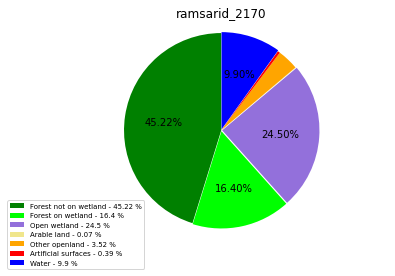
ramsarid_2171
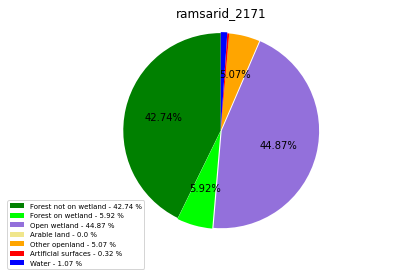
ramsarid_2172
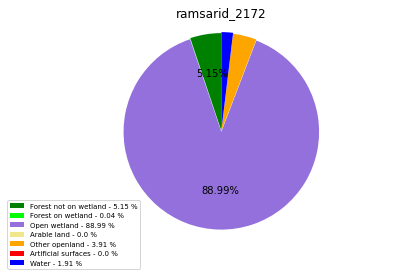
ramsarid_2173
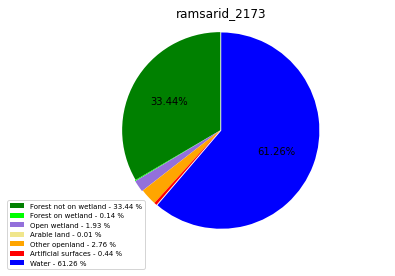
ramsarid_2174
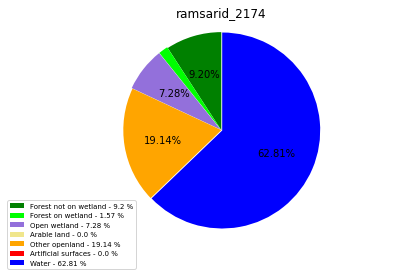
ramsarid_2175
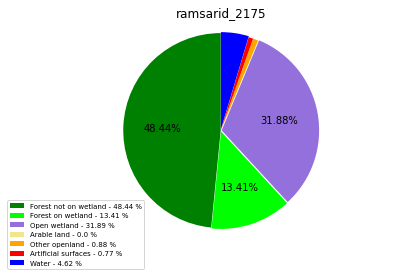
ramsarid_2176
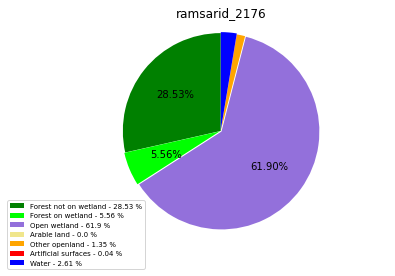
ramsarid_2177
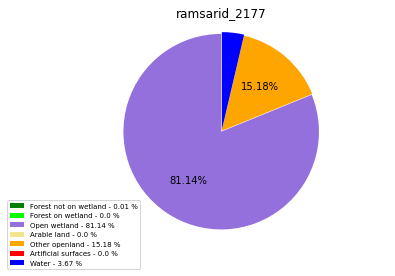
ramsarid_2178
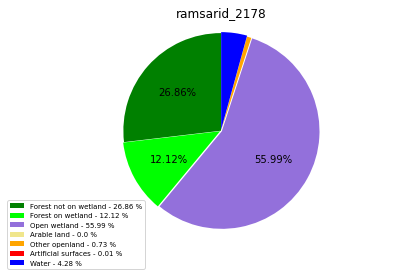
ramsarid_2179
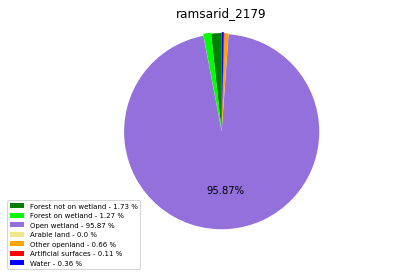
ramsarid_2180
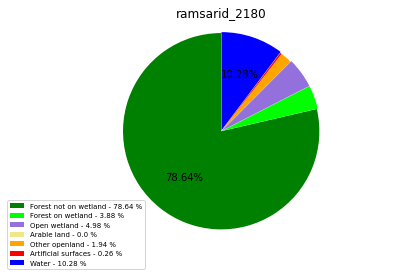
ramsarid_22
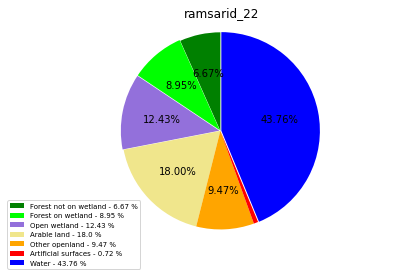
ramsarid_2260
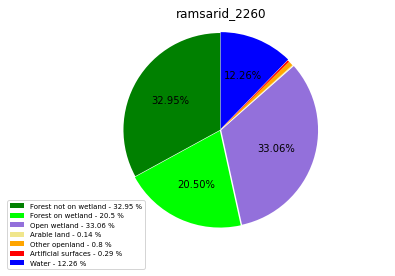
ramsarid_2265
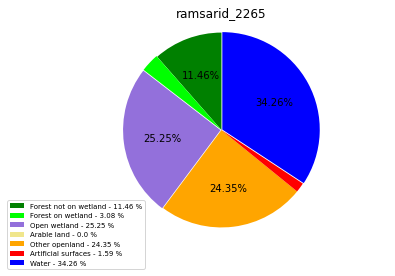
ramsarid_23
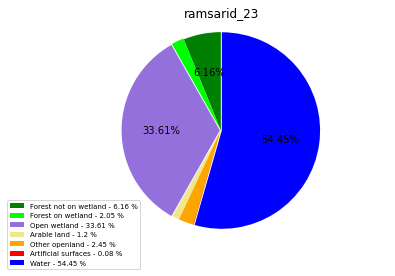
ramsarid_24
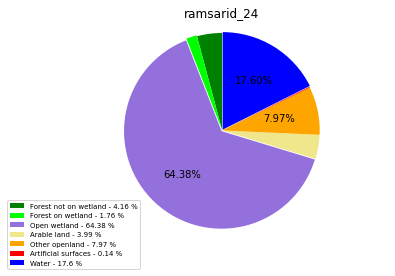
ramsarid_25
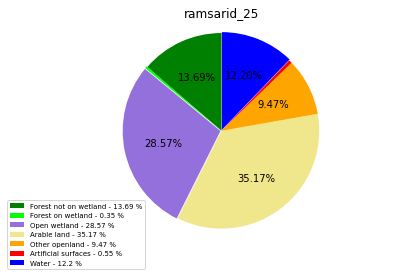
ramsarid_26
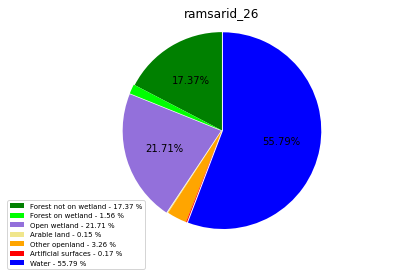
ramsarid_27
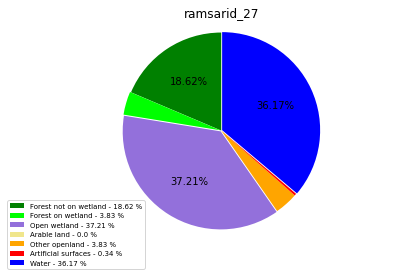
ramsarid_28
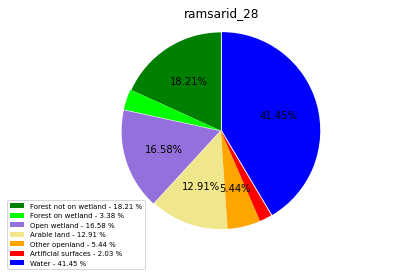
ramsarid_29
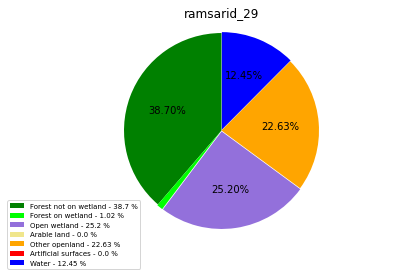
ramsarid_30
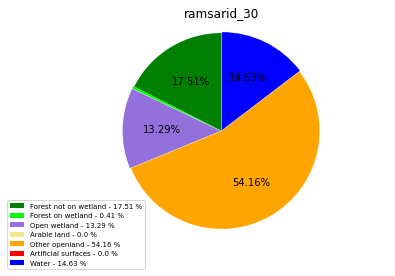
ramsarid_31
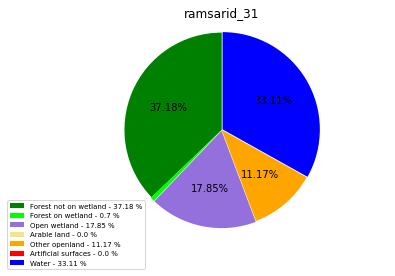
ramsarid_32
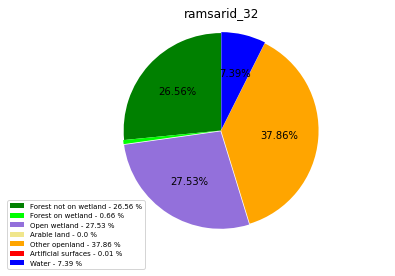
ramsarid_33
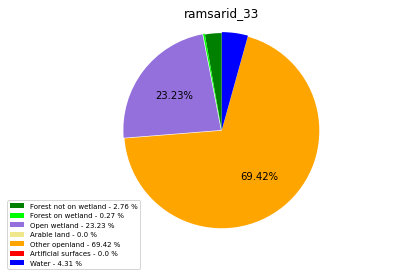
ramsarid_429
ramsarid_430
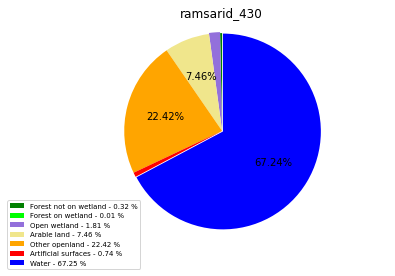
ramsarid_431
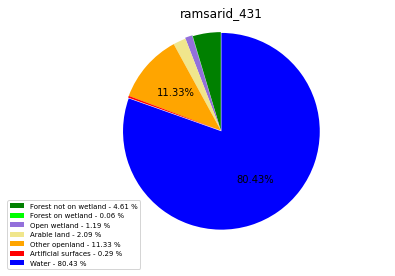
ramsarid_432
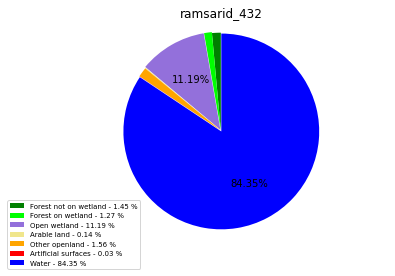
ramsarid_433
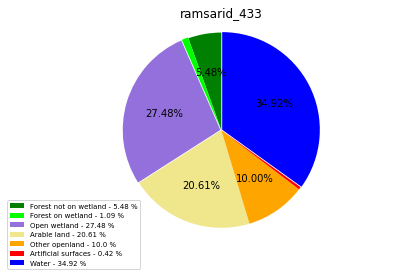
ramsarid_434
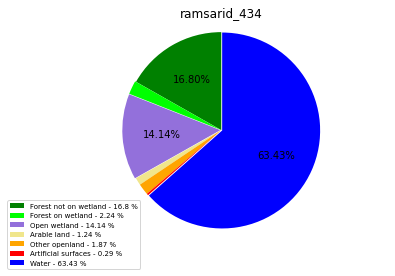
ramsarid_435
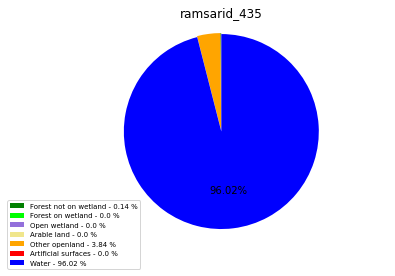
ramsarid_436
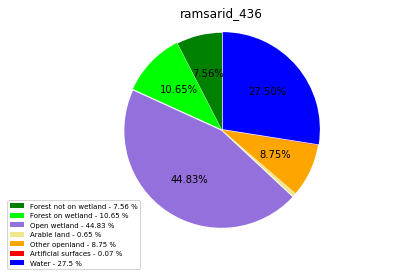
ramsarid_437
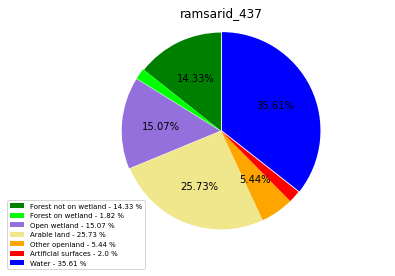
ramsarid_438
1.1.2. Open water and flooded vegetation extraction
1.1.2.1. Open water extraction
Layers: SAR intensity sentinel-1 image.

data source: Google Earth Engine
we use thresholding method to extract open water from S1 layer
First we move all layers to the crs: EPSG:4326
[109]:
%%bash
rm --force layers_reprj/*.tif
for file in layers/*.tif; do echo $file; filename=$(basename $file .tif);
gdalwarp -ot "Float32" -t_srs EPSG:4326 $file layers_reprj/${filename}_reprj.tif;done
layers/coherence.tif
Creating output file that is 15046P x 4166L.
Processing layers/coherence.tif [1/1] : 0Using internal nodata values (e.g. -3.4e+38) for image layers/coherence.tif.
Copying nodata values from source layers/coherence.tif to destination layers_reprj/coherence_reprj.tif.
...10...20...30...40...50...60...70...80...90...100 - done.
layers/S2.tif
Creating output file that is 3119P x 1352L.
Processing layers/S2.tif [1/1] : 0Using internal nodata values (e.g. -3.4e+38) for image layers/S2.tif.
Copying nodata values from source layers/S2.tif to destination layers_reprj/S2_reprj.tif.
...10...20...30...40...50...60...70...80...90...100 - done.
layers/SAR_VH.tif
Creating output file that is 3119P x 1352L.
Processing layers/SAR_VH.tif [1/1] : 0Using internal nodata values (e.g. -3.4e+38) for image layers/SAR_VH.tif.
Copying nodata values from source layers/SAR_VH.tif to destination layers_reprj/SAR_VH_reprj.tif.
...10...20...30...40...50...60...70...80...90...100 - done.
layers/slope.tif
Creating output file that is 1040P x 452L.
Processing layers/slope.tif [1/1] : 0Using internal nodata values (e.g. -3.4e+38) for image layers/slope.tif.
Copying nodata values from source layers/slope.tif to destination layers_reprj/slope_reprj.tif.
...10...20...30...40...50...60...70...80...90...100 - done.
[111]:
%%bash
#applying nodata value equal to zero
for file in layers/*.tif; do echo $file;
gdal_edit.py -a_nodata -3.39999995214436425e+38 $file;done
layers/coherence.tif
layers/S2.tif
layers/SAR_VH.tif
layers/slope.tif
[1]:
%%bash
rm --force crop/*.tif
for file in layers/*.tif; do echo $file;
filename=$(basename $file .tif)
gdalwarp -overwrite -te 14.6714649 58.2914991 14.9516494 58.4129513 -tr 0.000269494585236 -0.000269494585236 $file crop/${filename}_crop.tif
done
layers/coherence.tif
Creating output file that is 1040P x 451L.
Processing layers/coherence.tif [1/1] : 0Using internal nodata values (e.g. -3.4e+38) for image layers/coherence.tif.
Copying nodata values from source layers/coherence.tif to destination crop/coherence_crop.tif.
...10...20...30...40...50...60...70...80...90...100 - done.
layers/S2.tif
Creating output file that is 1040P x 451L.
Processing layers/S2.tif [1/1] : 0Using internal nodata values (e.g. -3.4e+38) for image layers/S2.tif.
Copying nodata values from source layers/S2.tif to destination crop/S2_crop.tif.
...10...20...30...40...50...60...70...80...90...100 - done.
layers/SAR_VH.tif
Creating output file that is 1040P x 451L.
Processing layers/SAR_VH.tif [1/1] : 0Using internal nodata values (e.g. -3.4e+38) for image layers/SAR_VH.tif.
Copying nodata values from source layers/SAR_VH.tif to destination crop/SAR_VH_crop.tif.
...10...20...30...40...50...60...70...80...90...100 - done.
layers/slope.tif
Creating output file that is 1040P x 451L.
Processing layers/slope.tif [1/1] : 0Using internal nodata values (e.g. -3.4e+38) for image layers/slope.tif.
Copying nodata values from source layers/slope.tif to destination crop/slope_crop.tif.
...10...20...30...40...50...60...70...80...90...100 - done.
we maskout values greater than -23
[ ]:
%%bash
pkgetmask -max -23 -data 2 -nodata 0 -ot Byte -i crop/SAR_VH_crop.tif -o crop/water.tif;
gdalwarp -crop_to_cutline -cutline rms/ramsarid_23.gpkg crop/water.tif crop/wat.tif;
1.1.2.2. Flooded vegetation extraction by using unsupervised Kmean classification
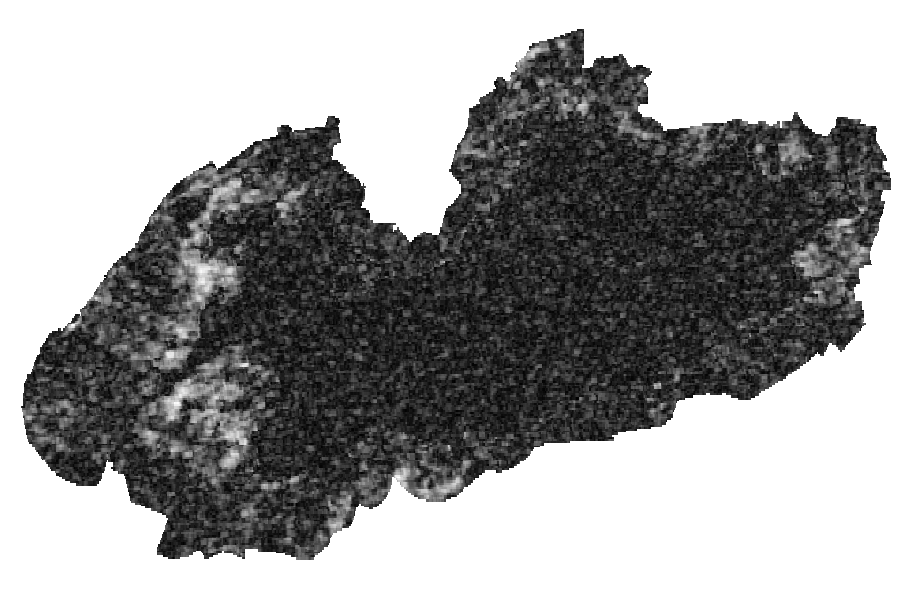
Layers: 1. Coherence layer ( left picture) 2. slope layer (generated from SRTM DEM (right picture)

orfeo kmean clustering toolbox could not handle nan values and alsso give us warning for nodatavalue=-3.39999995214436425e+38 by the following code we solve this two problems
[142]:
%%bash
#crop the coherence, SAR, and slope .tif files by our wetland shape of interest to exclude the nan values
gdalwarp -crop_to_cutline -overwrite -srcnodata -3.39999995214436425e+38 -dstnodata -999 -cutline rms/ramsarid_23.gpkg crop/slope_crop.tif crop/slope_crop_ready.tif;
gdalwarp -crop_to_cutline -overwrite -srcnodata -3.39999995214436425e+38 -dstnodata -999 -cutline rms/ramsarid_23.gpkg crop/coherence_crop.tif crop/coherence_crop_ready.tif
Creating output file that is 792P x 254L.
Processing crop/slope_crop.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
Creating output file that is 792P x 254L.
Processing crop/coherence_crop.tif [1/1] : 0...10...20...30...40...50...60...70...80...90...100 - done.
We want to build stack of 2 layers before doing unsupervised classification: SAR coherence and SRTM DEM
[143]:
%%bash
# stck the tif
gdalbuildvrt -overwrite -separate crop/stack.vrt crop/coherence_crop_ready.tif crop/slope_crop_ready.tif
gdalinfo -mm crop/stack.vrt
0...10...20...30...40...50...60...70...80...90...100 - done.
Driver: VRT/Virtual Raster
Files: crop/stack.vrt
crop/coherence_crop_ready.tif
crop/slope_crop_ready.tif
Size is 792, 254
Coordinate System is:
GEOGCS["WGS 84",
DATUM["WGS_1984",
SPHEROID["WGS 84",6378137,298.257223563,
AUTHORITY["EPSG","7030"]],
AUTHORITY["EPSG","6326"]],
PRIMEM["Greenwich",0],
UNIT["degree",0.0174532925199433],
AUTHORITY["EPSG","4326"]]
Origin = (14.707577174421624,58.387888303573057)
Pixel Size = (0.000269494585236,-0.000269494585236)
Corner Coordinates:
Upper Left ( 14.7075772, 58.3878883) ( 14d42'27.28"E, 58d23'16.40"N)
Lower Left ( 14.7075772, 58.3194367) ( 14d42'27.28"E, 58d19' 9.97"N)
Upper Right ( 14.9210169, 58.3878883) ( 14d55'15.66"E, 58d23'16.40"N)
Lower Right ( 14.9210169, 58.3194367) ( 14d55'15.66"E, 58d19' 9.97"N)
Center ( 14.8142970, 58.3536625) ( 14d48'51.47"E, 58d21'13.18"N)
Band 1 Block=128x128 Type=Float32, ColorInterp=Undefined
Computed Min/Max=0.000,0.741
NoData Value=-999
Band 2 Block=128x128 Type=Float32, ColorInterp=Undefined
Computed Min/Max=0.000,15.575
NoData Value=-999
[150]:
%%bash
# unsupervised classification
~/OTB/superbuild_install/bin/otbcli_KMeansClassification -in crop/stack.vrt -ts 1000 -nc 15 -maxit 1000 -out crop/ClassificationFilterOutput.tif uint8
2021-06-02 21:53:21 (INFO) KMeansClassification: Default RAM limit for OTB is 256 MB
2021-06-02 21:53:21 (INFO) KMeansClassification: GDAL maximum cache size is 398 MB
2021-06-02 21:53:21 (INFO) KMeansClassification: OTB will use at most 2 threads
2021-06-02 21:53:21 (INFO) KMeansClassification: Elevation management: setting default height above ellipsoid to 0 meters
(0s)
2021-06-02 21:53:21 (INFO) KMeansClassification: add field in the layer ...
2021-06-02 21:53:21 (INFO) KMeansClassification: Polygon Class Statistics...
2021-06-02 21:53:21 (INFO) KMeansClassification: Elevation management: setting default height above ellipsoid to 0 meters
2021-06-02 21:53:21 (INFO): Estimated memory for full processing: 2.99329MB (avail.: 256 MB), optimal image partitioning: 1 blocks
2021-06-02 21:53:21 (INFO): Estimation will be performed in 1 blocks of 792x254 pixels
Analyze polygons...: 100% [**************************************************] (0s)
2021-06-02 21:53:21 (INFO) KMeansClassification: 1000 is the maximum sample size that will be used.
2021-06-02 21:53:21 (INFO) KMeansClassification: Sample selection...
2021-06-02 21:53:21 (INFO) KMeansClassification: Elevation management: setting default height above ellipsoid to 0 meters
2021-06-02 21:53:21 (INFO) KMeansClassification: Sampling strategy : set a constant number of samples for all classes
2021-06-02 21:53:21 (INFO) KMeansClassification: Sampling rates : className requiredSamples totalSamples rate
0 1000 201168 0.00497097
2021-06-02 21:53:21 (INFO): Estimated memory for full processing: 2.99329MB (avail.: 256 MB), optimal image partitioning: 1 blocks
2021-06-02 21:53:21 (INFO): Estimation will be performed in 2 blocks of 448x254 pixels
Selecting positions with periodic sampler...: 100% [**************************************************] (0s)
2021-06-02 21:53:22 (INFO): Estimated memory for full processing: 4.52808MB (avail.: 256 MB), optimal image partitioning: 1 blocks
2021-06-02 21:53:22 (INFO): Estimation will be performed in 1 blocks of 792x254 pixels
Extracting sample values...: 100% [**************************************************] (0s)
2021-06-02 21:53:22 (INFO) KMeansClassification: Compute Images second order statistics...
2021-06-02 21:53:22 (INFO): Estimated memory for full processing: 4.52808MB (avail.: 256 MB), optimal image partitioning: 1 blocks
2021-06-02 21:53:22 (INFO): Estimation will be performed in 1 blocks of 792x254 pixels
Processing Image (1/1): 100% [**************************************************] (0s)
2021-06-02 21:53:22 (INFO) KMeansClassification: image statistics file: crop/ClassificationFilterOutput.tif_imgstats.xml
2021-06-02 21:53:22 (INFO) KMeansClassification: Model training...
2021-06-02 21:53:22 (INFO) KMeansClassification: Reading vector file 1/1
2021-06-02 21:53:22 (INFO) KMeansClassification: Computing model file : crop/ClassificationFilterOutput.tif_model.txt
Training model...: 100% [**************************************************] (0s)
Validation...: 100% [**************************************************] (0s)
2021-06-02 21:53:22 (INFO) KMeansClassification: Training performances:
2021-06-02 21:53:22 (INFO) KMeansClassification: Contingency table: reference labels (rows) vs. produced labels (cols)
ContingencyTableCalculator (0x558686a52230)
labels 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14
0 437 64 21 16 43 59 58 59 70 31 41 26 9 51 15
2021-06-02 21:53:22 (INFO) KMeansClassification: output model: crop/ClassificationFilterOutput.tif_model.txt
2021-06-02 21:53:22 (INFO) KMeansClassification: Performs a classification of the input image...
2021-06-02 21:53:22 (INFO) KMeansClassification: Loading model
2021-06-02 21:53:22 (INFO) KMeansClassification: Model loaded
2021-06-02 21:53:22 (INFO) KMeansClassification: Input image normalization activated.
2021-06-02 21:53:22 (INFO) KMeansClassification: mean used: [-430.072, -429.681]
2021-06-02 21:53:22 (INFO) KMeansClassification: standard deviation used: [494.746, 495.087]
2021-06-02 21:53:22 (INFO) KMeansClassification: Final clean-up ...
2021-06-02 21:53:23 (INFO): Estimated memory for full processing: 8.81551MB (avail.: 256 MB), optimal image partitioning: 1 blocks
2021-06-02 21:53:23 (INFO): File crop/ClassificationFilterOutput.tif will be written in 1 blocks of 792x254 pixels
Writing crop/ClassificationFilterOutput.tif...: 100% [**************************************************] (0s)
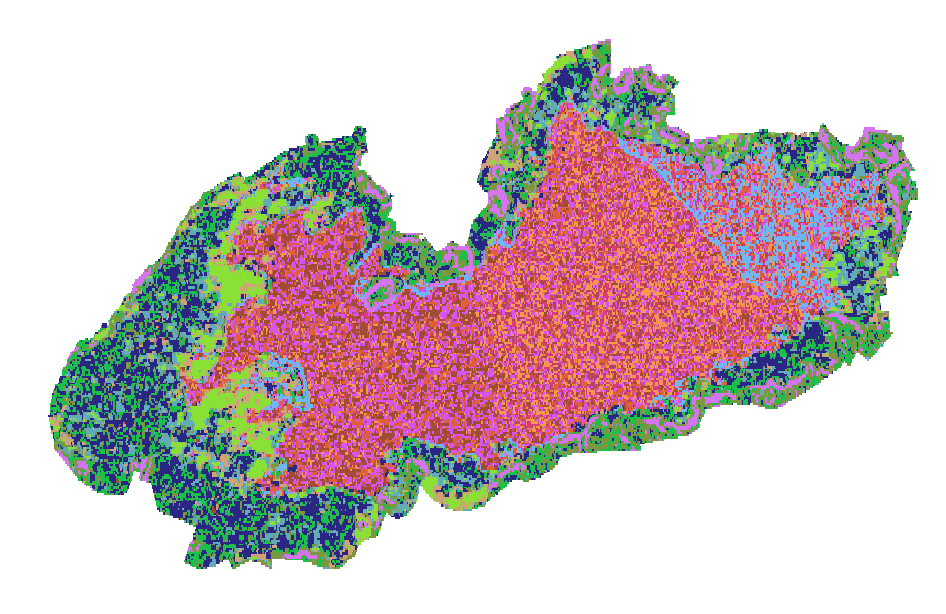
In the following code we select the cluster with highest coherence (cluster n. 12)
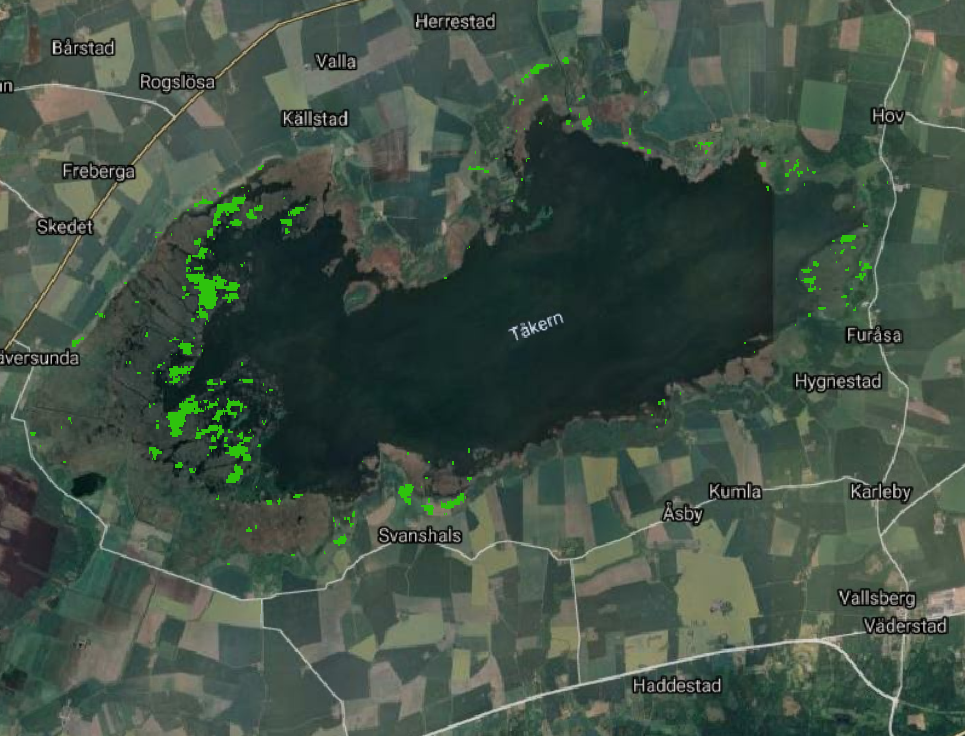
[138]:
%%bash
pkgetmask -max 12 -min 12 -data 1 -nodata 0 -ot Byte -i crop/ClassificationFilterOutput.tif -o crop/veg.tif
0...10...20...30...40...50...60...70...80...90...100 - done.
Now we want to calculate number of water pixels and flooded Vegetation
[151]:
%%bash
#convert .tif to .txt
gdal_translate -of XYZ crop/wat.tif crop/wat.txt
gdal_translate -of XYZ crop/veg.tif crop/veg.txt
Input file size is 792, 254
0...10...20...30...40...50...60...70...80...90...100 - done.
Input file size is 792, 254
0...10...20...30...40...50...60...70...80...90...100 - done.
[1]:
%%bash
#calculating number of pixels
awk '{print$3}' crop/wat.txt | grep 2 | wc -l > crop/final.txt;
awk '{print$3}' crop/veg.txt | grep 1 | wc -l >> crop/final.txt;
cat crop/final.txt
58524
1220
[155]:
!jupyter nbconvert my_project.ipynb --to html
[NbConvertApp] Converting notebook my_project.ipynb to html
[NbConvertApp] Writing 2321636 bytes to my_project.html