Python data analysis
Saverio Mancino
Open the notebook lesson
Open the lesson using the CL
cd /media/sf_LVM_shared/my_SE_data/exercise
jupyter-lab Python_data_analysis_SM.ipynb
Lesson Overview
This lesson is designed to provide a thorough understanding of data handling using Python. You will learn how to work with numerical data using Numpy and handle tabular data using Pandas. The lesson includes:
Detailed explanations of key concepts;
Python code examples in executable cells;
Exercise boxes with challenges to test your understanding.
Objectives
By the end of this lesson, you should be able to:
Knowing the Jupyter enviroment for python programming;
Understand the basics of Numpy for numerical operations
Create and manipulate Numpy arrays
Understand the fundamentals of Pandas, including Series and DataFrames
Import, clean, and analyze data using Pandas
Apply advanced data handling techniques such as grouping, merging, and pivoting
Solve practical exercises to reinforce your learning
Table of Contents
Jupyter Environment for Python Programming
GitHub and Code Repository Versioning Systems
Introduction
Numpy Basics
Pandas Basics
Exercises
1 - Environments for Python Programming
Python Kernel
The Python kernel is the component that executes your Python code within a Jupyter Notebook. When you run a cell, the kernel processes the code and returns the output. Also, the kernel maintains the state of your session, including variable definitions, imported modules, and function declarations. This means that cells can depend on code executed in previous cells. Infact, Jupyter provides also the options to restart or interrupt the kernel. This is useful for clearing the workspace or stopping long-running processes without having to close the entire notebook.
Jupyter Notebook
Jupyter is an open-source, locally managed, web coding application for project development.

A Jupyter notebook has two components: a front-end web page and a back-end kernel. The front-end web page allows data scientists to enter programming code or text into rectangular ‘cells’. The browser then passes the code to the back-end kernel, which executes it and returns the results. Its characterised by:
- Interactive Computing:Jupyter Notebooks let you run code in an interactive, cell-by-cell manner. This enables iterative development and immediate feedback, which is especially useful when testing new ideas or debugging.
- Integrated Documentation:Combine rich text (using Markdown), live code, and visualizations in a single document. This integration supports reproducible research and thorough documentation of your data analysis workflow.
- Multi-Language Support:Although originally designed for Python and Bash, Jupyter supports many programming languages (e.g., R, Julia) through the use of kernels. This flexibility allows you to work with multiple languages in one environment.
- Ease of Sharing:Notebooks can be easily shared and converted into different formats (HTML, PDF, slides), making it simple to disseminate your work among colleagues or publish it online.
Google Colab
Google Colab is a cloud-based Jupyter Notebook environment developed by Google.

It allows users to write and execute Python code in an interactive notebook format without requiring any local setup. Google Colab is particularly useful for Python beginners, researchers, and data scientists who want a hassle-free environment for coding, data analysis, and machine learning experiments.
- No Installation Required:Colab runs entirely in the cloud, so there’s no need to install Python, Jupyter, or any dependencies.
- Free GPU and TPU Access:Google provides free access to GPUs and TPUs, making it a great choice for machine learning and data science projects.
- Integration with Google Drive:You can save and load files directly from Google Drive, making it easy to store and share your work.
- Collaboration:Multiple users can edit and run the same notebook simultaneously, similar to Google Docs.
- Pre-installed Libraries:Colab comes already with popular Python libraries such as NumPy, Pandas, TensorFlow, and Matplotlib pre-installed.
Using Google Colab:
Access Colab: Open Google Colab in your browser.
Create a New Notebook: Click on “New Notebook” to start coding in Python.
Upload Files: Use files.upload() from google.colab to upload datasets.
2 - GitHub and Code Repository Versioning Systems
In modern software development and data science projects, version control systems are essential for maintaining code integrity, tracking changes, and enabling collaboration. This subchapter provides an overview of GitHub, along with other code repository and versioning systems commonly used in the field.
What is Version Control?
Version control is a system that records changes to a file or set of files over time. This allows you to:
Revert to Previous Versions: Easily roll back to earlier iterations if errors or issues arise.
Collaborate Efficiently: Multiple developers can work on the same project simultaneously without overwriting each other’s work.
Track Changes: Maintain a detailed history of modifications, including who made the changes and why.
Git and Its Ecosystem
Git: Git is a distributed version control system that lets every developer maintain a complete local copy of the project history. Its branching and merging features facilitate experimentation and efficient collaboration.
Repositories: A repository (or repo) is the storage space for your project’s files along with their version history. Repositories can be local (on your machine) or hosted remotely.
GitHub

What is GitHub? GitHub is a web-based platform built around Git. It provides an intuitive interface for hosting, managing, and collaborating on Git repositories.
Key Features:
Pull Requests: Enable developers to propose changes, review code collaboratively, and merge updates into the main project after thorough discussion.
Issue Tracking & Project Management: Built-in tools to manage bugs, track feature requests, and plan project workflows.
Continuous Integration (CI): Integration with CI/CD tools automates testing and deployment, ensuring that code changes meet quality standards before they are merged.
Community & Open Source: GitHub hosts millions of open-source projects, making it a vibrant community for sharing and contributing to software development.
There are many other popular platforms like GitLab, Bitbucket etc., wich offers similar functionalities to GitHub with a different emphasis on other features.
3 - Introduction
In today’s world, data is at the heart of decision-making across industries: from scientific research to finance, healthcare and social media. This lesson is designed to equip you with essential skills in data handling using Python, focusing on two of its most powerful libraries: Numpy and Pandas.
The Importance of Data Handling
With the exponential growth in the volume of data generated every day, having robust tools and methodologies to handle, clean, and analyze data is more crucial than ever. Thats why an effective data management and analysis can uncover insights that drive strategic decisions. Expecially in conducting scientific research the ability to interpret data accurately is invaluable. Automated data handling tools use is spreading to minimizes human error and increases the speed of data processing. By leveraging Python’s libraries, you can streamline tasks that would otherwise be time-consuming if done manually. Its important to be comfortable with manipulating data in Python but also prepared to tackle more complex data challenges, in this sense mastering Numpy and Pandas lays the groundwork for more advanced topics in data science, including machine learning, statistical analysis, and data visualization.
Bash vs Python: Memory Usage and Use Cases
Bash is efficient for:
Handling large text files (e.g.,
awk,sed,grep);Processing streams without loading data into memory;
Automating workflows and integrating different programs;
Dealing with high-latency remote server/cluster.
However, Bash has limitations in:
Complex data structures (e.g., arrays, dictionaries);
Advanced mathematical operations;
Readability and debugging;
When to Use Python
Python language is preferred for:
Complex data manipulations (e.g., NumPy, Pandas);
Machine learning and data analysis;
Scripts requiring structured programming and logic;
However, Python differently from bash, loads all data into memory, which can be inefficient for very large files compared to streaming in Bash.
Language Snippets
One of the most powerfull tecnique consinst in using small code snippets of Python in Bash or vice versa. Switching code language with snippets it’s like switching gears while driving a car.
Bash as the first gear: Ideal for quick tasks, file manipulation, and streaming large datasets without loading them entirely.
Python as the fifth gear: Powerfull, best for complex calculations, structured data manipulations, and advanced analysis and modeling.
Mastering the switch between Bash and Python snippets can significantly optimize workflows and memory usage.
What You Will Learn
Fundamentals of Numpy: Learn how to create and manipulate multi-dimensional arrays, perform vectorized operations, and understand how these methods offer significant performance improvements over traditional Python loops.
Pandas for Structured Data: Discover how to create and manage data using Pandas DataFrames and Series. You’ll learn methods for cleaning, transforming, and summarizing data, enabling you to work efficiently with large datasets.
Advanced Techniques: Delve into more sophisticated operations such as merging datasets, grouping data for aggregate analysis, and creating pivot tables to reorganize and summarize complex data structures.
Hands-On Practice: This lesson incorporates practical exercises and code examples, giving you the opportunity to apply theoretical knowledge to real-world data scenarios. Each exercise is designed to reinforce your learning and build your confidence in using Python for data handling.
Jupyterlab setup and libraries retrieving [Just for UBUNTU 24.04]
To run this lesson properly on osboxes.org you need to have a clean install of jupyterlab. So you need to run this command lines on your terminal:
pipx uninstall jupyterlab
pipx install --system-site-packages jupyterlab
Then you need to install python libraries using the apt always in the command line, in this way:
sudo apt install python3-numpy
sudo apt install python3-pandas
Data Retrieving
Before proceeding with the lesson, run this code to download to the virtual machine the files that will be used in these python lessons.
OSGEOLIVE16 proceding
If you’re working on osgeolive16 you can procede with the installation of gdown, to directly download from a google drive repo:
!pip install gdown
then procede to move and extract files from the downloaded files.zip, after that removing the archive:
import gdown
!mkdir -p /media/sf_LVM_shared/my_SE_data/exercise/files
file_url = 'https://drive.google.com/uc?export=download&id=1J54Xpk-qHnz7yuIYSqOgVQy6_QpYhLVv'
output_path = '/media/sf_LVM_shared/my_SE_data/exercise/files/file.zip'
gdown.download(file_url, output_path, quiet=True)
!unzip /media/sf_LVM_shared/my_SE_data/exercise/files/file.zip -d /media/sf_LVM_shared/my_SE_data/exercise/files
!rm /media/sf_LVM_shared/my_SE_data/exercise/files/file.zip
all files will be available in:
/media/sf_LVM_shared/my_SE_data/exercise/files
UBUNTU24 proceding
If you’re working on Ubuntu 24.04 you can procede directly with:
!mkdir -p /media/sf_LVM_shared/my_SE_data/exercise/files
then download manually your files from:
https://drive.google.com/uc?export=download&id=1J54Xpk-qHnz7yuIYSqOgVQy6_QpYhLVv
then procede to move and extract files from the downloaded files.zip, after that removing the archive:
!mv /home/osboxes/Downloads/files.zip /media/sf_LVM_shared/my_SE_data/exercise/files/
!unzip /media/sf_LVM_shared/my_SE_data/exercise/files/file.zip -d /media/sf_LVM_shared/my_SE_data/exercise/files
!rm /media/sf_LVM_shared/my_SE_data/exercise/files/file.zip
all files will be available in:
/media/sf_LVM_shared/my_SE_data/exercise/files
4 - Numpy Basics

Creating Numpy arrays;
Basic arithmetic and vectorized operations;
Indexing, slicing, and reshaping arrays;
import.as instead.numpy as np[3]:
# Importing necessary libraries
import numpy as np
import pandas as pd
from PIL import Image
If numpy import doesn’t work properly the following bash lines should be run
[4]:
# !pip show numpy pandas | grep Version:
[5]:
# !pip uninstall -y numpy
[6]:
# !pip install numpy==1.23.0
np.name_of_function.Arrays
Arrays in Python
In Python, arrays can be implemented in different ways:
Lists: Python’s built-in lists can store different data types but are not optimized for numerical computations.
NumPy Arrays: NumPy provides specialized arrays that are more memory-efficient and allow fast mathematical operations.
Numpy arrays
np.array([]).[7]:
np.array([1,2,3,4,5,6,7,8,9])
[7]:
array([1, 2, 3, 4, 5, 6, 7, 8, 9])
[8]:
arr1 = np.array([1,2,3,4])
arr2 = np.array([3,4,5,6])
np.add(arr1, arr2)
[8]:
array([ 4, 6, 8, 10])
We can also add arrays using the following convenient notation:
[9]:
arr1 + arr2
[9]:
array([ 4, 6, 8, 10])
[ start : end : stride ].[10]:
arr = np.array([0,1,2,3,4,5,6,7,8,9,10,11,12,13,14,15])
print(arr[5]) # show 5° position
print(arr[5:]) # start from 5° position
print(arr[:5]) # end on 5° position
print(arr[::2]) # Reading step set as 2
5
[ 5 6 7 8 9 10 11 12 13 14 15]
[0 1 2 3 4]
[ 0 2 4 6 8 10 12 14]
[11]:
arr = np.array([0,1,2,3,4,5,6,7,8,9,10,11,12,13,14,15])
arr[-2:] # show the first 2 elements from the last position
[11]:
array([14, 15])
[12]:
# We can create a Numpy array
a = np.array([1, 2, 3, 4, 5])
print('Original array:', a)
# We can manipulate it with some basic arithmetic operations
# adding elements
b = a + 1
print('After adding one:', b)
# or subtracting elements
slice_a = a[1:4]
print('Slice (indexes 1 to 3):', slice_a)
Original array: [1 2 3 4 5]
After adding one: [2 3 4 5 6]
Slice (indexes 1 to 3): [2 3 4]
[ ].[ ] represent the first dimension and the innermost [ ] contains the last dimension.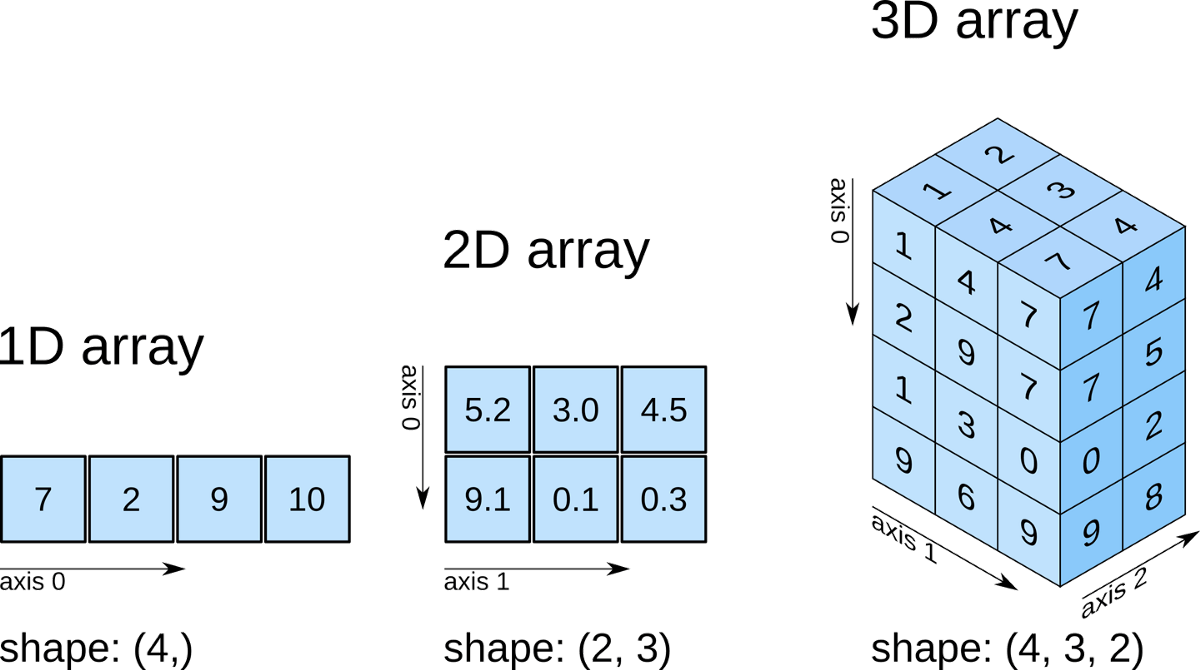
Now we declare a 2D array with shape (1, 9). In this case, the nested (double) square brackets [[ ]] indicates the array is 2-dimensional.
[13]:
np.array([[1,2,3,4,5,6,7,8,9]])
[13]:
array([[1, 2, 3, 4, 5, 6, 7, 8, 9]])
To visualise the shape (dimensions) of a numpy array we can add the suffix .shape to an array expression or variable containing a numpy array.
[14]:
arr1 = np.array([1,2,3,4,5,6,7,8,9]) # 1D array
arr2 = np.array([[1,2,3,4,5,6,7,8,9]]) # 2D array
arr3 = np.array([[1],[2],[3],[4],[5],[6],[7],[8],[9]]) # 2D array
arr4 = np.array([1,2,3]) # 1D array
print(f"\
arr1 shape: {arr1.shape} \
arr2 shape: {arr2.shape} \
arr3 shape: {arr3.shape} \
arr4 shape: {arr4.shape} \
")
arr1 shape: (9,) arr2 shape: (1, 9) arr3 shape: (9, 1) arr4 shape: (3,)
Unsigned Integers |
|
|---|---|
bits |
alias |
8 bits |
uint8 |
16 bits |
uint16 |
32 bits |
uint32 |
64 bits |
uint64 |
Signed Integers |
|
|---|---|
bits |
alias |
8 bits |
int8 |
16 bits |
int16 |
32 bits |
int32 |
64 bits |
int64 |
Floats |
|
|---|---|
bits |
alias |
32 bits |
float32 |
64 bits |
float64 |
We can look up the type of an array by using the .dtype suffix.
[15]:
arr = np.ones((10,10,10))
# In this way we created a 10x10x10 matrix populated only by '1'
# arr
print (f"shape: {arr.shape}")
print (f"type: {arr.dtype}")
print (f"weight: {round((arr.nbytes / 1024),2)} kB")
arr_bool = arr.astype(bool)
print (f"type: {arr_bool.dtype}")
print (f"weight: {round((arr_bool.nbytes / 1024),2)} kB")
shape: (10, 10, 10)
type: float64
weight: 7.81 kB
type: bool
weight: 0.98 kB
bool.True or False.[16]:
arr = np.array([True, False, True]) # declaring a 1D bool array
print("bool array:", arr)
print("array shape: ", arr.shape)
print("array type: ", arr.dtype)
bool array: [ True False True]
array shape: (3,)
array type: bool
Numpy Operations
Arithmetic Operations: Numpy supports element-wise operations, allowing you to add, subtract, multiply, or divide arrays directly.
Slicing and Indexing: Similar to Python lists, arrays can be sliced using the
[start:stop]syntax.
Let’s explore further operations such as reshaping and broadcasting.
and and or.* (and), and + (or).Note: Here the ``*`` and ``+`` symbols are not performing multiplication and addition as with numerical arrays. Numpy detects the type of the arrays involved in the operation and changes the behaviour of these operators.
[17]:
arr1 = np.array([True, True, False, False])
arr2 = np.array([True, False, True, False])
# two way to use AND operator
print ("AND operator")
print(np.logical_and(arr1, arr2))
print(arr1 * arr2)
# two way to use OR operator
print ("OR operator")
print(np.logical_or(arr1, arr2))
print(arr1 + arr2)
AND operator
[ True False False False]
[ True False False False]
OR operator
[ True True True False]
[ True True True False]
[18]:
arr = np.array([1, 3, 5, 1, 6, 3, 1, 5, 7, 1])
print(arr == 1)
print(arr > 2)
[ True False False True False False True False False True]
[False True True False True True False True True False]
False values from a numeric array.True values in the mask array.[19]:
arr = np.array([1,2,3,4,5,6,7,8,9])
mask = np.array([True,False,True,False,True,False,True,False,True])
var= arr[mask]
NumPy automatically ‘stretches’ the smaller array across the larger one so that their shapes become compatible for element-wise operations. This becomes widely useful in data pre-processing and in pixel-wise filtering procedures.[20]:
# Example: Broadcasting
matrix = np.array([[1, 2, 3], [4, 5, 6]])
print('matrix:')
print(matrix)
print(' ')
vector = np.array([10, 20, 30])
print('vector:')
print(vector)
print(' ')
# Adding a vector to each row of the matrix using broadcasting
broadcast_sum = matrix + vector
print('Broadcasting addition:')
print(broadcast_sum)
print(' ')
# Subtracting a vector to each row of the matrix using broadcasting
broadcast_sub = abs(matrix - vector)
print('Broadcasting subtraction:')
print(broadcast_sub)
print(' ')
matrix:
[[1 2 3]
[4 5 6]]
vector:
[10 20 30]
Broadcasting addition:
[[11 22 33]
[14 25 36]]
Broadcasting subtraction:
[[ 9 18 27]
[ 6 15 24]]
Scalar and Matrix Products
np.dot.
[21]:
# Section 4: Scalar and Matrix Products
# Dot product of two vectors
a = np.array([2, 7, 1])
b = np.array([8, 2, 8])
print(f"first vector:\n{a}\n\nsecond vector:\n{b}\n")
dot_product = np.dot(a, b)
print('Dot product:', dot_product)
first vector:
[2 7 1]
second vector:
[8 2 8]
Dot product: 38
While, to perform a matrix multiplications we can use the @ operator as a shorthand for np.matmul.
[22]:
# Matrix multiplication using the @ operator
matrix_a = np.array([[1, 2], [3, 4]])
matrix_b = np.array([[5, 6], [7, 8]])
print(f"first matrix:\n{matrix_a}\n\nsecond matrix:\n{matrix_b}\n")
matrix_product = matrix_a @ matrix_b
print(f"Matrix product:\n{matrix_product}")
first matrix:
[[1 2]
[3 4]]
second matrix:
[[5 6]
[7 8]]
Matrix product:
[[19 22]
[43 50]]
Data Cleaning and Processing with NumPy
NumPy.np.nan) and potential outliers.[23]:
# Create sample data
data = np.array([1.0, 1.5, 1.8, 1.9, 1.9, 121.5, 2.0, 2.1, 2.2, 2.2, 2.3, 2.5, 2.9, 3.1, 3.5, np.nan, 4.2, 100.0, 3.8, np.nan, 2.7])
print(f"Original Data:\n{data}")
Original Data:
[ 1. 1.5 1.8 1.9 1.9 121.5 2. 2.1 2.2 2.2 2.3 2.5
2.9 3.1 3.5 nan 4.2 100. 3.8 nan 2.7]
Numpy tools.np.isnan, crucial for understanding the extent and location of missing data before applying any cleaning techniques.[24]:
# Identifying missing values (Binary NaN mask)
missing_mask = np.isnan(data)
print(f"Missing Data Mask:{missing_mask}\n")
# Counting missing values
num_missing = np.sum(missing_mask)
print(f"Number of missing values:{num_missing}")
Missing Data Mask:[False False False False False False False False False False False False
False False False True False False False True False]
Number of missing values:2
using
np.nan_to_numto replacenp.nanwith 0,using
np.wherealong withnp.nanmeanto replace missing values with the mean of the non-missing data.
[25]:
# Replacing np.nan with "0" using np.nan_to_num
data_filled = np.nan_to_num(data, nan=0.0)
print(f"Data after replacing missing values with 0:\n{data_filled}\n")
# Replace np.nan with the rounded mean of non-missing values using np.nanman
mean_value = round(np.nanmean(data),2)
data_mean_filled = np.where(np.isnan(data), mean_value, data)
print(f"The data mean value is:\n{mean_value}")
print(f"Data after replacing missing values with the mean:\n{data_mean_filled}")
Data after replacing missing values with 0:
[ 1. 1.5 1.8 1.9 1.9 121.5 2. 2.1 2.2 2.2 2.3 2.5
2.9 3.1 3.5 0. 4.2 100. 3.8 0. 2.7]
The data mean value is:
13.85
Data after replacing missing values with the mean:
[ 1. 1.5 1.8 1.9 1.9 121.5 2. 2.1 2.2 2.2
2.3 2.5 2.9 3.1 3.5 13.85 4.2 100. 3.8 13.85
2.7 ]
[26]:
# Definition of an outlier threshold (e.g., values >= 20 are considered outliers)
threshold = 20
filtered_data = data[data < threshold]
print(f"Data after filtering out outliers (values >= 20):\n{filtered_data}")
Data after filtering out outliers (values >= 20):
[1. 1.5 1.8 1.9 1.9 2. 2.1 2.2 2.2 2.3 2.5 2.9 3.1 3.5 4.2 3.8 2.7]
np.sort to order the data and np.unique to find distinct values; along with the calculation of the basic aggregate statistics like the sum (np.sum) and mean (np.mean) of the cleaned data.[27]:
# Sort the data after filling missing values
sorted_data = np.sort(filtered_data)
print(f"Sorted Data:\n{sorted_data}")
# Identify unique values
unique_values = np.unique(filtered_data)
print(f"Unique Values:\n'{unique_values}")
# Calculate aggregate statistics
data_sum = round(np.sum(filtered_data),2)
data_mean = round(np.mean(filtered_data),2)
data_std = round(np.std(filtered_data),2)
print('\n\
Sum:', data_sum, '\n\
Mean:', data_mean, '\n\
Std:', data_std)
Sorted Data:
[1. 1.5 1.8 1.9 1.9 2. 2.1 2.2 2.2 2.3 2.5 2.7 2.9 3.1 3.5 3.8 4.2]
Unique Values:
'[1. 1.5 1.8 1.9 2. 2.1 2.2 2.3 2.5 2.7 2.9 3.1 3.5 3.8 4.2]
Sum: 41.6
Mean: 2.45
Std: 0.81
np.where for conditional data transformations with np.clip to limit the values within a specified range, which is often useful for handling extreme values.[28]:
# Conditional processing example: data gain by multiply values > than 3 by 100
condition = filtered_data > 3
data_processed = np.where(condition, filtered_data * 100, filtered_data)
print(f"Data after conditional processing (values < 3 multiplied by 100):\n{data_processed}")
# Data ranging using np.clip to limit values to a range (e.g., 1 to 3)
data_clipped = np.clip(data_filled, 1, 3)
print(f"Data after clipping values to the range 0-3:\n'{data_clipped}")
Data after conditional processing (values < 3 multiplied by 100):
[ 1. 1.5 1.8 1.9 1.9 2. 2.1 2.2 2.2 2.3 2.5 2.9
310. 350. 420. 380. 2.7]
Data after clipping values to the range 0-3:
'[1. 1.5 1.8 1.9 1.9 3. 2. 2.1 2.2 2.2 2.3 2.5 2.9 3. 3. 1. 3. 3.
3. 1. 2.7]
Numpy tools, along with many others, are essential for manipulating and processing numerical data, expecially when those are (often) very large.5 - Pandas Basics
Now we will explore Pandas, a powerful open-source library built on top of NumPy, widely used for data manipulation and analysis. It introduces DataFrames and Series, which are highly efficient for working with tabular data.
In this section, we cover:
Basic functionality of the library,
DataFrames and tools for exploration,
Creating and manipulating Pandas Series and DataFrames,
Data cleaning and standardization functionalities,
Data importing and manipulatng from structured file formats,
Basic data exploration and summary statistics
In Pandas we can create two main kind of data structure:
Pandas Series (1D labeld array);
Pandas DataFrame (2D labeled data structure).
head(), tail(), and info(), which help understand the dataset quickly.[29]:
# Creating a Pandas Series
series = pd.Series([1, "a", 3, "b", 5])
print(f"Pandas Series:\n\
{series}")
Pandas Series:
0 1
1 a
2 3
3 b
4 5
dtype: object
[30]:
print("Same Pandas series seen with the enhanced Pandas data visualizzation:")
series
Same Pandas series seen with the enhanced Pandas data visualizzation:
[30]:
0 1
1 a
2 3
3 b
4 5
dtype: object
Pandas Series
enumerate(series.items()) to extrapolate positions and values of some cells, also posing some particular conditions.series.apply to apply a lambda x function, as a small anonymous function inside another function for retrieving data.isinstance() function, that returns True if the specified object is of the specified type, otherwise False.[31]:
# Basic exploration and analysis of the Pd series
print(f"Series total length:\n{len(series)}\n")
print("Series type and position analysis:")
for i, (index, value) in enumerate(series.items()):
print(f"Position: {index}, Value: {value}, Type: {type(value)}")
# Localization of specific cells
# Strings localization
strings = series.apply(lambda x: isinstance(x, str))
print(f"\nNumber of strings cells:{len(series[strings])}")
print("\nPosition of strings cells:")
print(series[strings])
# Int localization
integers = series.apply(lambda x: isinstance(x, int))
print(f"\nNumber of integers cells:{len(series[integers])}")
print("\nPosition of integers cells:")
print(series[integers])
Series total length:
5
Series type and position analysis:
Position: 0, Value: 1, Type: <class 'int'>
Position: 1, Value: a, Type: <class 'str'>
Position: 2, Value: 3, Type: <class 'int'>
Position: 3, Value: b, Type: <class 'str'>
Position: 4, Value: 5, Type: <class 'int'>
Number of strings cells:2
Position of strings cells:
1 a
3 b
dtype: object
Number of integers cells:3
Position of integers cells:
0 1
2 3
4 5
dtype: object
Pandas DataFrame
starting from a dictionary,
populated row by row,
by the combination of other dataframes or data structures.
It offers a variety of tools for exploring data, such as .head(), .tail(),.sort_index, .sort_value and .info(), which help understand the dataset quickly.”
[32]:
# Creating a DataFrame and Basic Exploration
# Create a DataFrame from a dictionary
data = {
'Station': ['S01', 'S02', 'S03', 'S04','S05','S06'], #col1
'AverageTemp (T°C)': [25.5, 26.1, 27.3, 25.8, 25.1, 24.9,], #col2
'AverageUmidity (%)': [58, 67, 70, 61, 59, 55] #col3
}
df = pd.DataFrame(data)
print(f"Pandas dataframe:\n {df}")
Pandas dataframe:
Station AverageTemp (T°C) AverageUmidity (%)
0 S01 25.5 58
1 S02 26.1 67
2 S03 27.3 70
3 S04 25.8 61
4 S05 25.1 59
5 S06 24.9 55
[33]:
print("Same Pandas dataframe seen with the enhanced Pandas data visualizzation:")
df
Same Pandas dataframe seen with the enhanced Pandas data visualizzation:
[33]:
| Station | AverageTemp (T°C) | AverageUmidity (%) | |
|---|---|---|---|
| 0 | S01 | 25.5 | 58 |
| 1 | S02 | 26.1 | 67 |
| 2 | S03 | 27.3 | 70 |
| 3 | S04 | 25.8 | 61 |
| 4 | S05 | 25.1 | 59 |
| 5 | S06 | 24.9 | 55 |
[34]:
# Basic exploration and analysis of the Pd Dataframe
# General basic information about the DataFrame
print("DataFrame Info:\n")
print(f"{df.info()}\n")
# Display the first few rows
print(f"DataFrame Head:\n{df.head()}\n")
# Display the last few rows
print(f"DataFrame tail:\n{df.tail()}\n")
# Display the dataframe sorted by descending values
print(f"DataFrame sorted by AverageTemp (T°C) values:\n{df.sort_values(by=['AverageTemp (T°C)'],ascending=False)}\n")
# Display the dataframe sorted by descending indexes
print(f"DataFrame sorted by descending indexes:\n{df.sort_index(ascending=False)}")
DataFrame Info:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 6 entries, 0 to 5
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Station 6 non-null object
1 AverageTemp (T°C) 6 non-null float64
2 AverageUmidity (%) 6 non-null int64
dtypes: float64(1), int64(1), object(1)
memory usage: 276.0+ bytes
None
DataFrame Head:
Station AverageTemp (T°C) AverageUmidity (%)
0 S01 25.5 58
1 S02 26.1 67
2 S03 27.3 70
3 S04 25.8 61
4 S05 25.1 59
DataFrame tail:
Station AverageTemp (T°C) AverageUmidity (%)
1 S02 26.1 67
2 S03 27.3 70
3 S04 25.8 61
4 S05 25.1 59
5 S06 24.9 55
DataFrame sorted by AverageTemp (T°C) values:
Station AverageTemp (T°C) AverageUmidity (%)
2 S03 27.3 70
1 S02 26.1 67
3 S04 25.8 61
0 S01 25.5 58
4 S05 25.1 59
5 S06 24.9 55
DataFrame sorted by descending indexes:
Station AverageTemp (T°C) AverageUmidity (%)
5 S06 24.9 55
4 S05 25.1 59
3 S04 25.8 61
2 S03 27.3 70
1 S02 26.1 67
0 S01 25.5 58
In this way we can easly strip the DataFrame in all its “informational direction” and sample its contents, detecting any issues or crucial information.
adding new columns,
renaming columns,
performing basic arithmetic operations.
[35]:
# Adding a new column to the DataFrame
df['NewColumn'] = [True, True, True, True, False, True]
print(f"DataFrame with a new column:\n{df}\n")
# Renaming a column
df = df.rename(columns={'NewColumn': 'Online'})
print(f"DataFrame with renamed column:\n{df}\n")
DataFrame with a new column:
Station AverageTemp (T°C) AverageUmidity (%) NewColumn
0 S01 25.5 58 True
1 S02 26.1 67 True
2 S03 27.3 70 True
3 S04 25.8 61 True
4 S05 25.1 59 False
5 S06 24.9 55 True
DataFrame with renamed column:
Station AverageTemp (T°C) AverageUmidity (%) Online
0 S01 25.5 58 True
1 S02 26.1 67 True
2 S03 27.3 70 True
3 S04 25.8 61 True
4 S05 25.1 59 False
5 S06 24.9 55 True
dropna()andfillna()for missing data filling,.str.title()and.astype()for data standardizaion.
[36]:
# Create a DataFrame with missing values and inconsistent strings
raw_data = {
'Station': ['S01', 's02', np.nan, 's03', np.nan],
'Measurement(T°C)': [15, np.nan, 15.33, 10.2, np.nan],
'City': ['New York', 'los angeles', 'Chicago', np.nan, np.nan],
}
df_raw = pd.DataFrame(raw_data)
print(f"Original DataFrame with Missing/Inconsistent Data:\n\
{df_raw}\n")
# Keep only the rows with at least 2 non-NA values and create a copy.
df_filtered = df_raw.dropna(thresh=2).copy()
print(f"Original DataFrame with at least 2 non NaN values:\n\
{df_filtered}\n")
# Calculate mean from original dataset
mean_measurement = df_raw['Measurement(T°C)'].mean()
# Filling DataFrame missing numerical values, with the mean of that column, and forcing values to int
df_filtered.loc[:, 'Measurement(T°C)'] = df_filtered['Measurement(T°C)'].fillna(mean_measurement).astype(int)
# Filling missing categorical values with a placeholder,
df_filtered.loc[:, 'Station'] = df_filtered['Station'].fillna('Unknown')
df_filtered.loc[:, 'City'] = df_filtered['City'].fillna('Unknown')
# Standardize string upper/lower cases
df_filtered.loc[:, 'Station'] = df_filtered['Station'].str.title()
df_filtered.loc[:, 'City'] = df_filtered['City'].str.title()
print(f"Cleaned and Standardized DataFrame:\n{df_filtered}")
Original DataFrame with Missing/Inconsistent Data:
Station Measurement(T°C) City
0 S01 15.00 New York
1 s02 NaN los angeles
2 NaN 15.33 Chicago
3 s03 10.20 NaN
4 NaN NaN NaN
Original DataFrame with at least 2 non NaN values:
Station Measurement(T°C) City
0 S01 15.00 New York
1 s02 NaN los angeles
2 NaN 15.33 Chicago
3 s03 10.20 NaN
Cleaned and Standardized DataFrame:
Station Measurement(T°C) City
0 S01 15.0 New York
1 S02 13.0 Los Angeles
2 Unknown 15.0 Chicago
3 S03 10.0 Unknown
Data import
pd.read_csv and perform basic manipulations on it.[37]:
# Data retrieved from https://www.dati.gov.it/view-dataset/dataset?id=9bacb31d-1b49-4841-b87e-8a442e133aa8
# Reading a CSV file
df_imported = pd.read_csv('files/Dati_Meteo_Giornalieri_Stazione _Matera.csv')
print("\
Daily weather data taken from the weather station installed on top of the Matera Town Hall.\n\
The data will be collected daily at 12:00 noon, unless otherwise unforeseen impediments.\n\
Legend:\n \
- TM=average temperature,\n \
- UM=average humidity,\n \
- VVM=average wind speed m/s,\n \
- PRE=precipitation mm,\n \
- RSM=average solar radiation,\n \
- PATM=average atmospheric pressure,\n \
- QA PS PM=air quality fine particulate matter,\n \
- QA PG PM=coarse dust air quality.\
")
print("\n Imported DataFrame from CSV:")
df_imported
Daily weather data taken from the weather station installed on top of the Matera Town Hall.
The data will be collected daily at 12:00 noon, unless otherwise unforeseen impediments.
Legend:
- TM=average temperature,
- UM=average humidity,
- VVM=average wind speed m/s,
- PRE=precipitation mm,
- RSM=average solar radiation,
- PATM=average atmospheric pressure,
- QA PS PM=air quality fine particulate matter,
- QA PG PM=coarse dust air quality.
Imported DataFrame from CSV:
[37]:
| DATA | TM | UM | VVM | PRE | RSM | PATM | QA PS PM | QA PG PM | ORA | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 03/12/2021 | 7,420 | 95,300 | 2,750 | 0,250 | 24,6 | 951 | 0,500 | 0,150 | 12:00 |
| 1 | 04/12/2021 | 9,490 | 72,600 | 3,590 | 0,000 | 35,9 | 957 | 0,530 | 0,000 | 12:00 |
| 2 | 05/12/2021 | 11,900 | 79,900 | 3,970 | 0,000 | 38,4 | 952 | 0,040 | 0,010 | 12:00 |
| 3 | 06/12/2021 | 7,400 | 85,800 | 2,120 | 0,000 | 185,0 | 947 | 0,000 | 0,000 | 12:00 |
| 4 | 07/12/2021 | 6,480 | 72,900 | 4,980 | 0,000 | 320,0 | 952 | 0,140 | 0,100 | 12:00 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 200 | 05/08/2022 | 33,500 | 21,200 | 1,670 | 0,000 | 794,0 | 960 | 1,020 | 0,900 | 12:00 |
| 201 | 06/08/2022 | ND | ND | ND | ND | ND | ND | ND | ND | 12:00 |
| 202 | 07/08/2022 | ND | ND | ND | ND | ND | ND | ND | ND | 12:00 |
| 203 | 08/08/2022 | ND | ND | ND | ND | ND | ND | ND | ND | 12:00 |
| 204 | 10/08/2022 | 27,100 | 53,500 | 4,400 | 0,000 | 533,0 | 961,0 | 1,370 | 1,500 | 12:00 |
205 rows × 10 columns
.describe().[38]:
# Basic Data Exploration and Summary Statistics
print(f"Summary Statistics for Imported DataFrame:\n{df_imported.describe()}\n")
Summary Statistics for Imported DataFrame:
DATA TM UM VVM PRE RSM PATM QA PS PM QA PG PM ORA
count 205 205 205 205 205 205 205 205 205 205
unique 205 157 172 161 6 173 37 136 48 2
top 03/12/2021 ND ND ND 0,000 ND ND ND 0,000 12:00
freq 1 15 15 15 185 15 15 15 50 204
.str.contains('').any() for localizing any problematic string, value or symbol.replace() for replacing that with a functional one.[39]:
print(f"PRE column before any cleaning:\n{df_imported['PRE'].tail()}\n")
print(f"TM column before any cleaning:\n{df_imported['TM'].tail()}\n")
# Data cleaning
# Control to Replace any 'ND' with the computation friendly 'NaN',
# For precipitation data
if df_imported['PRE'].astype(str).str.contains('ND').any():
df_imported['PRE'] = df_imported['PRE'].replace('ND', np.nan)
# For temperature data
if df_imported['TM'].astype(str).str.contains('ND').any():
df_imported['TM'] = df_imported['TM'].replace('ND', np.nan)
# Control to replace any comma with dot to standardize decimal separator
# For precipitation data
if df_imported['PRE'].astype(str).str.contains(',').any():
df_imported['PRE'] = df_imported['PRE'].str.replace(',', '.')
# For temperature data
if df_imported['TM'].astype(str).str.contains(',').any():
df_imported['TM'] = df_imported['TM'].str.replace(',', '.')
# Convert the column to numeric, coercing errors to NaN
# For precipitation data
df_imported['PRE'] = pd.to_numeric(df_imported['PRE'], errors='coerce')
# For temperature data
df_imported['TM'] = pd.to_numeric(df_imported['TM'], errors='coerce')
# Verify cleaning
print(f"PRE column after cleaning:\n{df_imported['PRE'].tail()}\n")
print(f"TM column after cleaning:\n{df_imported['TM'].tail()}\n")
PRE column before any cleaning:
200 0,000
201 ND
202 ND
203 ND
204 0,000
Name: PRE, dtype: object
TM column before any cleaning:
200 33,500
201 ND
202 ND
203 ND
204 27,100
Name: TM, dtype: object
PRE column after cleaning:
200 0.0
201 NaN
202 NaN
203 NaN
204 0.0
Name: PRE, dtype: float64
TM column after cleaning:
200 33.5
201 NaN
202 NaN
203 NaN
204 27.1
Name: TM, dtype: float64
Then we use functions like describe(), max(), min(), mean() and median() to obtain summary statistics and gain insights into the data.
[40]:
# Summary statistics on the cleaned columns
# Calculate mean and median for the precipitation and average temperature column
max_precipitation = df_imported['PRE'].max()
min_precipitation = df_imported['PRE'].min()
mean_precipitation = round(df_imported['PRE'].mean(),2)
median_precipitation = round(df_imported['PRE'].median(),2)
max_temperature = df_imported['TM'].max()
min_temperature = df_imported['TM'].min()
mean_temperature = round(df_imported['TM'].mean(),2)
median_temperature = round(df_imported['TM'].median(),2)
print(f"\
----------------- PRECIPITATION --------------------------\n\
Max Precipitation : {max_precipitation} mm\n\
Min Precipitation : {min_precipitation} mm\n\
Mean Precipitation : {mean_precipitation} mm\n\
Median Precipitation : {mean_precipitation} mm\n\
----------------- TEMPERATURE --------------------------\n\
Max Temperature : {max_temperature} °C\n\
Min Temperature : {min_temperature} °C\n\
Mean Temperature : {mean_temperature} °C\n\
Median Temperature : {mean_temperature} °C\n\
")
----------------- PRECIPITATION --------------------------
Max Precipitation : 1.2 mm
Min Precipitation : 0.0 mm
Mean Precipitation : 0.01 mm
Median Precipitation : 0.01 mm
----------------- TEMPERATURE --------------------------
Max Temperature : 34.9 °C
Min Temperature : 1.27 °C
Mean Temperature : 16.96 °C
Median Temperature : 16.96 °C
We can use functions like value_counts() an .groupby() to get some deep specific statistics between different values present in the DataFrame.
[41]:
# Checking the number of events that exceeded 1 mm of Precipitation
print(f"\
Number of Precipitation events that exceede 1mm :\
\n\
{((df_imported['PRE']>1).value_counts())}\
\n\
")
df_imported['TM'] = pd.to_numeric(df_imported['TM'], errors='coerce')
print(f"\
Total rain mm for days having T<5°C:\
\n\
(T°C) (mm)\
\n\
{df_imported[df_imported['TM'] < 5].groupby('TM')['PRE'].sum()}\
")
Number of Precipitation events that exceede 1mm :
PRE
False 204
True 1
Name: count, dtype: int64
Total rain mm for days having T<5°C:
(T°C) (mm)
TM
1.27 0.4
2.18 0.0
3.13 0.0
3.79 0.0
3.83 0.0
3.99 0.0
4.34 0.0
4.52 0.0
4.71 0.0
Name: PRE, dtype: float64
.pivot_table() function is used to create spreadsheet-like pivot tables.[42]:
# Creating a climatic DataFrame as example
data = {
'Region': ['Puglia', 'Puglia', 'Puglia', 'Puglia',
'Basilicata', 'Basilicata', 'Basilicata', 'Basilicata',
'Molise', 'Molise', 'Molise', 'Molise'],
'Season': ['Inverno', 'Primavera', 'Estate', 'Autunno',
'Inverno', 'Primavera', 'Estate', 'Autunno',
'Inverno', 'Primavera', 'Estate', 'Autunno'],
'Mean_Temp(°C)': [8, 15, 27, 18, 5, 12, 25, 16, 3, 10, 24, 14],
'Mean_Prec(mm)': [80, 60, 20, 70, 100, 75, 30, 90, 120, 85, 40, 95],
'Mean_Umid(%)': [75, 65, 50, 70, 80, 70, 55, 75, 85, 75, 60, 78]
}
df_climatic = pd.DataFrame(data)
# Creating a pivot table to display average temperatures by region and season
print("Average temperatures by region and season:")
p_table = pd.pivot_table(df_climatic, values='Mean_Temp(°C)', index='Region', columns='Season', aggfunc='mean')
p_table
Average temperatures by region and season:
[42]:
| Season | Autunno | Estate | Inverno | Primavera |
|---|---|---|---|---|
| Region | ||||
| Basilicata | 16.0 | 25.0 | 5.0 | 12.0 |
| Molise | 14.0 | 24.0 | 3.0 | 10.0 |
| Puglia | 18.0 | 27.0 | 8.0 | 15.0 |
.to_csv() function, which allows you to export a DataFrame to a CSV file.[43]:
# Save DataFrame to CSV
df_climatic.to_csv(
'files/southern_regions_climate_data.csv', # name of the saved file
index=False, # Excludes the index column from the CSV
sep=';', # Set the separator (useful especially when we handle some natural language material inside some cells)
header=True # Keeps the header of the table in the CSV
)
print("File saved successfully.")
File saved successfully.
[44]:
import pandas as pd
#dataframe
df = pd.DataFrame([
[1, 23, 60],
[2, 22, 58]
], columns=['ID', 'Temp(°C)', 'Umid(%)'])
print("dataframe:\n ",df)
# array
arr = pd.Series([3, 28, 63])
df.loc[len(df)] = arr.values
print("\ndataframe after array appending as row:\n ",df)
dataframe:
ID Temp(°C) Umid(%)
0 1 23 60
1 2 22 58
dataframe after array appending as row:
ID Temp(°C) Umid(%)
0 1 23 60
1 2 22 58
2 3 28 63
[45]:
import pandas as pd
#dataframe
df = pd.DataFrame([
[1, 23, 60],
[2, 22, 58]
], columns=['ID', 'Temp(°C)', 'Umid(%)'])
print("dataframe:\n ",df)
# array
arr = pd.Series([0, 0.8])
df['Prec(mm)'] = arr.values
print("\ndataframe after array appending as column:\n ",df)
dataframe:
ID Temp(°C) Umid(%)
0 1 23 60
1 2 22 58
dataframe after array appending as column:
ID Temp(°C) Umid(%) Prec(mm)
0 1 23 60 0.0
1 2 22 58 0.8
[49]:
import pandas as pd
df = pd.DataFrame([
[1,25,38,0.00],
[2,26,37,0.18]
], columns=['ID','Temp(T°C)','Umid(%)','Prec(mm)'])
##manual user input into the dataframe
user_input = input ("insert new values into the table.\n\
use the format:\n\
[station ID number,Temperature in °C,Umidity as %,Precipitation as mm]\n\
with coma (',') as values separator:\n")
arr = pd.Series([x.strip() for x in user_input.split (',')])
df.loc[len(df)] = arr.values
df
insert new values into the table.
use the format:
[station ID number,Temperature in °C,Umidity as %,Precipitation as mm]
with coma (',') as values separator:
3, 40, 50, 0.20
[49]:
| ID | Temp(T°C) | Umid(%) | Prec(mm) | |
|---|---|---|---|---|
| 0 | 1 | 25 | 38 | 0.0 |
| 1 | 2 | 26 | 37 | 0.18 |
| 2 | 3 | 40 | 50 | 0.20 |
[50]:
import pandas as pd
df = pd.DataFrame([
[1,25,38,0.00],
[2,26,37,0.18]
], columns=['ID','Temp(T°C)','Umid(%)','Prec(mm)'])
label1 = 'station ID number'
label2 = 'Temperature in °C'
label3 = 'Umidity as %'
label4 = 'Precipitation as mm'
arr = []
for col in df.columns:
val = input(f"enter {col}:")
arr.append(val)
df.loc[len(df)] = arr
df
enter ID: 1
enter Temp(T°C): 23
enter Umid(%): 60
enter Prec(mm): 0.00
[50]:
| ID | Temp(T°C) | Umid(%) | Prec(mm) | |
|---|---|---|---|---|
| 0 | 1 | 25 | 38 | 0.0 |
| 1 | 2 | 26 | 37 | 0.18 |
| 2 | 1 | 23 | 60 | 0.00 |
6 - Exercises
Exercise: Integrative Data Analysis Challenge
Write code and document your process in Markdown cells.
Input
Dataset Import
Choose a publicly available CSV file, or
Use the provided dataset located at: >files/dataset.csv
Operations:
Data Cleaning with Pandas
Load the dataset into a Pandas DataFrame.
Explore the dataset using Pandas functions (e.g.,
.info(),.describe(),.head()).Handle missing values (e.g., forward fill, mean imputation, or removal),
Identify and filter out outliers if necessary,
Perform relevant data transformations (e.g., renaming columns, adjusting data types),
Compute aggregated statistics (e.g. mean, median, standard deviation etc.),
Extract meaningful statistics and create a pivot table using significant data,
Add a new empty column to the dataset.
Numerical Operations with NumPy
Normalize numerical columns if needed (e.g., adjusting types, handling decimal separators),
Utilize NumPy tools to process numerical values,
Populate the previously created empty column with calculated or transformed data.
Saving the Processed Data
Create a new directory for the exercise: >exercise/
Save the cleaned and processed dataset as: > exercise/result.csv
Output
A structured and cleaned dataset stored in: >exercise/result.csv
ready for further analysis in the next lesson.
[ ]:
## Do you exercise here:

