2.10. Ritwika Mukhopadhyay: Comparative Analysis of the prediction of AGB using Random Forest Regression, Support Vector Machine for Regression & FeedForward Neural Network
2.10.1. Project Description
The main objective was to predict forest aboveground biomass (AGB) with the metrics derived from airborne laser scanner (ALS) data. The field data for the sample plots were collected by Swedish National Forest Inventory (NFI) and a sub-area was selected from the entire forest region for this case study. Three models were compared on their efficiencies of AGB prediction - random forest regression (RFR), support vector machine for regression (SVR) and feed forward neural network (FF-NN). Finally the AGB predictions were extrapolated for the entire region using the wall-to-wall maps for the RFR.
2.10.2. Data Description
Field reference and ALS data were collected during the years 2015-19.
[1]:
# Import libraries
[1]:
import pandas as pd
import numpy as np
import rasterio
from rasterio import *
from rasterio.plot import show
from pyspatialml import Raster
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split,GridSearchCV
from sklearn.pipeline import Pipeline
from sklearn import metrics
from scipy.stats import pearsonr
import matplotlib.pyplot as plt
from sklearn.preprocessing import MinMaxScaler
from sklearn.svm import SVR
import os
import torch
import torch.nn as nn
import scipy
from sklearn.metrics import r2_score
from scipy import stats
from tabulate import tabulate
from sklearn.metrics import mean_absolute_error , mean_squared_error
plt.rcParams["figure.figsize"] = (10,6.5)
[ ]:
# Import dataset
[2]:
predictors = pd.read_csv("txt/NFI_all_T2.csv", sep= ",", index_col=False)
pd.set_option('display.max_columns',None)
# change column name
predictors = predictors.rename({'dev-magnitude':'devmagnitude', 'Elev P80':'p80', 'Canopy relief ratio':'crr'} , axis='columns')
predictors.head(10)
[2]:
| Taxar | TraktNr | PalslagNr | DelytaNr | IsPermanen | Ostkoordin | Nordkoordi | Lan | Lan_kort | Agoslag | Internatio | TorrviktSt | TorrviktGr | TorrviktRo | TorrviktAl | TotalAGB | id | datum | kommentar | flyghojd | flyghastig | overtackni | punkttathe | skannerid | skannerfab | skannermod | oppningsvi | pulsfrekve | skanningsf | uteffekt | square | Block | SkannDat | N | E | Las_Namn | Skanner2 | Region | LOV_AVL | NEAR_FID | NEAR_DIST | DataFile | FileTitle | Total return count | Total return count above 1.50 | Return 1 count above 1.50 | Return 2 count above 1.50 | Return 3 count above 1.50 | Return 4 count above 1.50 | Return 5 count above 1.50 | Return 6 count above 1.50 | Return 7 count above 1.50 | Return 8 count above 1.50 | Return 9 count above 1.50 | Other return count above 1.50 | Elev minimum | Elev maximum | Elev mean | Elev mode | Elev stddev | Elev variance | Elev CV | Elev IQ | Elev skewness | Elev kurtosis | Elev AAD | Elev MAD median | Elev MAD mode | Elev L1 | Elev L2 | Elev L3 | Elev L4 | Elev L CV | Elev L skewness | Elev L kurtosis | Elev P01 | Elev P05 | Elev P10 | Elev P20 | Elev P25 | Elev P30 | Elev P40 | Elev P50 | Elev P60 | Elev P70 | Elev P75 | p80 | Elev P90 | Elev P95 | Elev P99 | crr | Elev SQRT mean SQ | Elev CURT mean CUBE | Int minimum | Int maximum | Int mean | Int mode | Int stddev | Int variance | Int CV | Int IQ | Int skewness | Int kurtosis | Int AAD | Int L1 | Int L2 | Int L3 | Int L4 | Int L CV | Int L skewness | Int L kurtosis | Int P01 | Int P05 | Int P10 | Int P20 | Int P25 | Int P30 | Int P40 | Int P50 | Int P60 | Int P70 | Int P75 | Int P80 | Int P90 | Int P95 | Int P99 | Percentage first returns above 1.50 | Percentage all returns above 1.50 | (All returns above 1.50) / (Total first returns) * 100 | First returns above 1.50 | All returns above 1.50 | Percentage first returns above mean | Percentage first returns above mode | Percentage all returns above mean | Percentage all returns above mode | (All returns above mean) / (Total first returns) * 100 | (All returns above mode) / (Total first returns) * 100 | First returns above mean | First returns above mode | All returns above mean | All returns above mode | Total first returns | Total all returns | Profile area | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2017 | 5625 | 403 | 0 | 1 | 494081 | 6237269 | 10 | Blek | 1 | 1 | 65732.439490 | 19624.546900 | 27261.210090 | 112618.19650 | 112.618197 | 5313 | 20190406 | 3000 | 160 | 11 | 1.23 | 8238 | Leica | ALS80-HP | 35 | 278000 | 42 | 100 | 62350_4925_25 | 19A016 | 43561 | 6235000 | 492500 | 19A016_62350_4925_25.laz.laz | 0 | 0 | 2 | 3962 | 231 | D:\Projekt\ForestChangeHMB\LASdata\Skanning2_p... | 19A016_62350_4925_20175625403 | 811 | 440 | 358 | 75 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.51 | 17.270000 | 8.205409 | 2.010317 | 4.592859 | 21.094352 | 0.559735 | 8.515000 | 0.154795 | 1.664221 | 4.068788 | 4.300000 | 6.054683 | 8.205409 | 2.638618 | 0.137448 | -0.104379 | 0.321571 | 0.052091 | -0.039558 | 1.5600 | 2.029500 | 2.379000 | 3.278000 | 3.7500 | 4.224 | 6.342 | 8.065001 | 10.006000 | 11.785000 | 12.265000 | 12.850000 | 14.574000 | 15.250000 | 16.839298 | 0.424836 | 9.400806 | 10.277259 | 127 | 1622 | 382.668182 | 174.460317 | 248.884736 | 6.194361e+04 | 0.650393 | 234.00 | 2.060984 | 7.754890 | 177.008058 | 382.668182 | 120.554711 | 46.633416 | 25.590377 | 0.315037 | 0.386824 | 0.212272 | 147.119995 | 168.650009 | 185.400009 | 207.000000 | 216.00 | 229.000000 | 261.000000 | 305.5 | 351.000000 | 414.000000 | 450.00 | 495.000000 | 686.000061 | 958.000000 | 1302.879883 | 73.210634 | 54.254007 | 89.979550 | 358 | 440 | 40.899796 | 69.734151 | 26.140567 | 51.664612 | 43.353783 | 85.685072 | 200 | 341 | 212 | 419 | 489 | 811 | 26.578549 | |
| 1 | 2017 | 5627 | 103 | 0 | 1 | 476547 | 6239273 | 10 | Blek | 1 | 1 | 151893.416500 | 54522.253650 | 65733.906210 | 272149.57640 | 272.149576 | 5302 | 20190417 | 3000 | 160 | 11 | 1.23 | 8236 | Leica | ALS80-HP | 35 | 278000 | 38 | 100 | 62375_4750_25 | 19A016 | 43572 | 6237500 | 475000 | 19A016_62375_4750_25.laz.laz | 0 | 0 | 2 | 4131 | 727 | D:\Projekt\ForestChangeHMB\LASdata\Skanning2_p... | 19A016_62375_4750_20175627103 | 915 | 747 | 444 | 267 | 35 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1.51 | 20.660000 | 10.357416 | 6.981429 | 5.829134 | 33.978801 | 0.562798 | 11.200000 | 0.238884 | 1.580427 | 5.221311 | 4.590000 | 3.948572 | 10.357416 | 3.317782 | 0.237466 | -0.157953 | 0.320329 | 0.071574 | -0.047608 | 1.6846 | 2.190000 | 3.252000 | 4.762000 | 5.4800 | 6.000 | 7.080 | 8.740000 | 11.148000 | 15.642000 | 16.680000 | 17.459999 | 18.514000 | 19.020000 | 19.832399 | 0.462006 | 11.883156 | 13.030966 | 0 | 11226 | 496.734940 | 178.190476 | 636.854131 | 4.055832e+05 | 1.282080 | 347.00 | 12.983658 | 216.318892 | 266.425316 | 496.734940 | 186.958346 | 75.122169 | 52.911774 | 0.376374 | 0.401812 | 0.283014 | 126.000000 | 153.000000 | 171.000000 | 221.000000 | 241.00 | 271.000000 | 321.000000 | 389.0 | 461.000000 | 535.600098 | 588.00 | 658.599915 | 880.400146 | 1140.800171 | 1513.998901 | 94.067797 | 81.639344 | 158.262712 | 444 | 747 | 63.559322 | 74.788136 | 35.519126 | 50.491803 | 68.855932 | 97.881356 | 300 | 353 | 325 | 462 | 472 | 915 | 42.011135 | |
| 2 | 2017 | 5627 | 303 | 0 | 1 | 476282 | 6238964 | 10 | Blek | 1 | 1 | 117919.147600 | 47228.877970 | 54722.666020 | 219870.69160 | 219.870692 | 5302 | 20190417 | 3000 | 160 | 11 | 1.23 | 8236 | Leica | ALS80-HP | 35 | 278000 | 38 | 100 | 62375_4750_25 | 19A016 | 43572 | 6237500 | 475000 | 19A016_62375_4750_25.laz.laz | 0 | 0 | 2 | 4131 | 1036 | D:\Projekt\ForestChangeHMB\LASdata\Skanning2_p... | 19A016_62375_4750_20175627303 | 850 | 622 | 457 | 150 | 14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1.63 | 22.910000 | 15.888280 | 17.505555 | 3.493125 | 12.201924 | 0.219855 | 4.424999 | -0.698497 | 3.820452 | 2.754330 | 2.230000 | 2.330000 | 15.888280 | 1.941699 | -0.204725 | 0.247727 | 0.122210 | -0.105436 | 0.127583 | 6.9882 | 9.630000 | 11.464000 | 13.197999 | 13.8750 | 14.269 | 15.334 | 16.240000 | 17.120001 | 17.896999 | 18.299999 | 18.858000 | 20.077000 | 21.019001 | 22.095800 | 0.670032 | 16.267137 | 16.584793 | 0 | 11301 | 899.315113 | 179.380952 | 1332.791523 | 1.776333e+06 | 1.482007 | 749.75 | 6.679569 | 51.795140 | 569.119467 | 899.315113 | 404.862192 | 175.600160 | 134.662758 | 0.450189 | 0.433728 | 0.332614 | 150.000000 | 181.000000 | 211.599991 | 289.000000 | 343.00 | 399.200012 | 527.799927 | 661.5 | 837.400024 | 1013.000000 | 1092.75 | 1235.000000 | 1456.000244 | 1666.950195 | 11150.000000 | 96.210526 | 73.176471 | 130.947368 | 457 | 622 | 65.684211 | 45.052632 | 40.117647 | 26.588235 | 71.789474 | 47.578947 | 312 | 214 | 341 | 226 | 475 | 850 | 52.436067 | |
| 3 | 2017 | 5627 | 403 | 0 | 1 | 476268 | 6239247 | 10 | Blek | 1 | 1 | 5548.218087 | 6461.110215 | 3287.615349 | 15296.94365 | 15.296944 | 5302 | 20190417 | 3000 | 160 | 11 | 1.23 | 8236 | Leica | ALS80-HP | 35 | 278000 | 38 | 100 | 62375_4750_25 | 19A016 | 43572 | 6237500 | 475000 | 19A016_62375_4750_25.laz.laz | 0 | 0 | 2 | 4131 | 753 | D:\Projekt\ForestChangeHMB\LASdata\Skanning2_p... | 19A016_62375_4750_20175627403 | 506 | 133 | 133 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.53 | 4.290000 | 2.787895 | 2.318571 | 0.752301 | 0.565956 | 0.269845 | 1.110000 | 0.220942 | 2.063202 | 0.631294 | 0.570000 | 0.601429 | 2.787895 | 0.433357 | 0.023836 | 0.018626 | 0.155442 | 0.055002 | 0.042981 | 1.5728 | 1.696000 | 1.790000 | 1.998000 | 2.1900 | 2.310 | 2.524 | 2.810000 | 3.004000 | 3.220000 | 3.300000 | 3.412000 | 3.900000 | 4.128000 | 4.260400 | 0.455759 | 2.886877 | 2.979856 | 208 | 2211 | 960.150376 | 748.492063 | 504.494894 | 2.545151e+05 | 0.525433 | 573.00 | 0.897953 | 2.909312 | 398.217762 | 960.150376 | 276.435976 | 61.394064 | 34.786777 | 0.287909 | 0.222091 | 0.125840 | 259.320007 | 342.200012 | 404.799988 | 519.000000 | 623.00 | 679.000000 | 758.599976 | 834.0 | 921.199890 | 1048.200073 | 1196.00 | 1388.599854 | 1802.600220 | 2008.000000 | 2207.799805 | 28.913043 | 26.284585 | 28.913043 | 133 | 133 | 14.565217 | 20.000000 | 13.241107 | 18.181818 | 14.565217 | 20.000000 | 67 | 92 | 67 | 92 | 460 | 506 | 21.417354 | |
| 4 | 2019 | 5621 | 303 | 0 | 1 | 486266 | 6239102 | 10 | Blek | 1 | 1 | 129641.806700 | 32159.673020 | 53546.801660 | 215348.28140 | 215.348281 | 5308 | 20190406 | 3000 | 160 | 11 | 1.23 | 8238 | Leica | ALS80-HP | 35 | 278000 | 42 | 100 | 62375_4850_25 | 19A016 | 43561 | 6237500 | 485000 | 19A016_62375_4850_25.laz.laz | 0 | 0 | 2 | 4139 | 898 | D:\Projekt\ForestChangeHMB\LASdata\Skanning2_p... | 19A016_62375_4850_20195621303 | 828 | 386 | 309 | 73 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.93 | 27.760000 | 19.996762 | 24.480000 | 5.801991 | 33.663104 | 0.290147 | 6.467499 | -1.381331 | 4.499651 | 4.380333 | 3.139999 | 3.090000 | 19.996762 | 3.036699 | -0.877424 | 0.486469 | 0.151860 | -0.288940 | 0.160197 | 2.0740 | 6.145000 | 11.005000 | 16.719999 | 17.8225 | 18.510 | 20.270 | 21.430000 | 22.690001 | 23.889999 | 24.289999 | 24.680000 | 25.360001 | 26.160000 | 26.907499 | 0.699449 | 20.819375 | 21.356894 | 110 | 1671 | 310.647668 | 184.333333 | 213.085850 | 4.540558e+04 | 0.685941 | 106.75 | 2.970778 | 12.852277 | 128.962522 | 310.647668 | 88.865245 | 43.558159 | 31.810547 | 0.286064 | 0.490160 | 0.357964 | 142.649994 | 152.000000 | 169.000000 | 189.000000 | 199.25 | 207.000000 | 225.000000 | 252.0 | 271.000000 | 292.000000 | 306.00 | 333.000000 | 519.500000 | 793.000000 | 1155.549927 | 66.594828 | 46.618357 | 83.189655 | 309 | 386 | 47.629310 | 19.396552 | 28.502415 | 10.869565 | 50.862069 | 19.396552 | 221 | 90 | 236 | 90 | 464 | 828 | 34.031342 | |
| 5 | 2015 | 5625 | 103 | 0 | 1 | 491538 | 6239446 | 10 | Blek | 1 | 1 | 58841.982320 | 30217.391890 | 29542.145070 | 118601.51930 | 118.601519 | 5311 | 20190406 | 3000 | 160 | 11 | 1.23 | 8238 | Leica | ALS80-HP | 35 | 278000 | 42 | 100 | 62375_4900_25 | 19A016 | 43561 | 6237500 | 490000 | 19A016_62375_4900_25.laz.laz | 0 | 0 | 2 | 4143 | 554 | D:\Projekt\ForestChangeHMB\LASdata\Skanning2_p... | 19A016_62375_4900_20155625103 | 706 | 443 | 373 | 70 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.53 | 19.280001 | 13.715643 | 14.280000 | 2.795992 | 7.817574 | 0.203854 | 3.620000 | -0.610402 | 3.367664 | 2.198131 | 1.760000 | 1.699999 | 13.715643 | 1.559358 | -0.167807 | 0.212303 | 0.113692 | -0.107613 | 0.136148 | 6.2176 | 8.707001 | 9.952000 | 11.704000 | 12.0500 | 12.480 | 13.400 | 14.120000 | 14.612000 | 15.318000 | 15.670000 | 15.946000 | 17.271999 | 17.966999 | 18.803799 | 0.646707 | 13.997100 | 14.240406 | 144 | 13142 | 837.264108 | 144.000000 | 998.447381 | 9.968972e+05 | 1.192512 | 630.00 | 9.180494 | 103.197172 | 428.453801 | 837.264108 | 305.751836 | 99.859214 | 79.849615 | 0.365180 | 0.326602 | 0.261158 | 161.000000 | 216.399994 | 270.000000 | 364.000000 | 415.00 | 476.200012 | 585.000000 | 712.0 | 829.000000 | 967.000000 | 1045.00 | 1141.200073 | 1374.000000 | 1586.000000 | 1853.038086 | 82.705100 | 62.747875 | 98.226164 | 373 | 443 | 52.106430 | 44.345898 | 34.985836 | 29.036827 | 54.767184 | 45.454545 | 235 | 200 | 247 | 205 | 451 | 706 | 45.625354 | |
| 6 | 2015 | 5625 | 203 | 0 | 1 | 491548 | 6239171 | 10 | Blek | 1 | 1 | 0.000000 | 0.000000 | 0.000000 | 0.00000 | 0.000000 | 5311 | 20190406 | 3000 | 160 | 11 | 1.23 | 8238 | Leica | ALS80-HP | 35 | 278000 | 42 | 100 | 62375_4900_25 | 19A016 | 43561 | 6237500 | 490000 | 19A016_62375_4900_25.laz.laz | 0 | 0 | 2 | 4143 | 829 | D:\Projekt\ForestChangeHMB\LASdata\Skanning2_p... | 19A016_62375_4900_20155625203 | 429 | 34 | 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.51 | 2.780000 | 1.967353 | 1.510000 | 0.387121 | 0.149862 | 0.196772 | 0.470000 | 0.733778 | 2.283787 | 0.314844 | 0.235000 | 0.350000 | 1.967353 | 0.217282 | 0.048339 | 0.009820 | 0.110444 | 0.222471 | 0.045193 | 1.5166 | 1.530000 | 1.580000 | 1.612000 | 1.6425 | 1.686 | 1.784 | 1.860000 | 1.988000 | 2.045000 | 2.112500 | 2.386000 | 2.562000 | 2.720000 | 2.760200 | 0.360120 | 2.003979 | 2.041979 | 305 | 11658 | 3961.352941 | 1566.444444 | 4656.536624 | 2.168333e+07 | 1.175491 | 8273.00 | 1.010275 | 2.052198 | 4023.224913 | 3961.352941 | 2257.445633 | 870.321190 | 56.202227 | 0.569867 | 0.385534 | 0.024896 | 314.239990 | 338.850006 | 468.899994 | 631.000000 | 827.50 | 1097.199951 | 1617.000000 | 1692.5 | 1788.199951 | 1884.900024 | 9100.50 | 11488.000000 | 11572.000000 | 11602.099610 | 11658.000000 | 8.114558 | 7.925408 | 8.114558 | 34 | 34 | 3.818616 | 7.875895 | 3.729604 | 7.692308 | 3.818616 | 7.875895 | 16 | 33 | 16 | 33 | 419 | 429 | 13.492481 | |
| 7 | 2015 | 5625 | 403 | 0 | 1 | 491254 | 6239421 | 10 | Blek | 1 | 1 | 133254.384000 | 54100.341430 | 73852.081750 | 261206.80720 | 261.206807 | 5311 | 20190406 | 3000 | 160 | 11 | 1.23 | 8238 | Leica | ALS80-HP | 35 | 278000 | 42 | 100 | 62375_4900_25 | 19A016 | 43561 | 6237500 | 490000 | 19A016_62375_4900_25.laz.laz | 0 | 0 | 2 | 4143 | 579 | D:\Projekt\ForestChangeHMB\LASdata\Skanning2_p... | 19A016_62375_4900_20155625403 | 847 | 447 | 336 | 102 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.51 | 21.379999 | 13.186174 | 14.125872 | 5.084911 | 25.856324 | 0.385624 | 7.275000 | -0.634886 | 2.460292 | 4.178857 | 3.410000 | 3.515873 | 13.186174 | 2.859451 | -0.462348 | 0.208555 | 0.216852 | -0.161691 | 0.072935 | 1.7846 | 2.824000 | 5.029999 | 8.429999 | 9.9550 | 10.798 | 12.980 | 14.340000 | 15.246000 | 16.698000 | 17.230000 | 17.730000 | 19.062000 | 19.748001 | 20.718601 | 0.587628 | 14.130594 | 14.782048 | 127 | 11101 | 318.008949 | 127.000000 | 548.385200 | 3.007263e+05 | 1.724433 | 102.00 | 17.216338 | 335.506088 | 144.242442 | 318.008949 | 99.234267 | 60.864349 | 52.439225 | 0.312049 | 0.613340 | 0.528439 | 144.000000 | 161.000000 | 178.000000 | 195.000000 | 203.00 | 211.000015 | 225.000000 | 243.0 | 263.000000 | 288.000000 | 305.00 | 322.000000 | 424.000000 | 596.900208 | 1386.540894 | 76.190476 | 52.774498 | 101.360544 | 336 | 447 | 49.886621 | 45.351474 | 30.696576 | 27.744982 | 58.956916 | 53.287982 | 220 | 200 | 260 | 235 | 441 | 847 | 33.417988 | |
| 8 | 2016 | 5625 | 103 | 0 | 1 | 496555 | 6239468 | 10 | Blek | 1 | 1 | 67022.530310 | 16757.065520 | 27012.147500 | 110791.74330 | 110.791743 | 5315 | 20190406 | 3000 | 160 | 11 | 1.23 | 8238 | Leica | ALS80-HP | 35 | 278000 | 42 | 100 | 62375_4950_25 | 19A016 | 43561 | 6237500 | 495000 | 19A016_62375_4950_25.laz.laz | 0 | 0 | 2 | 4147 | 532 | D:\Projekt\ForestChangeHMB\LASdata\Skanning2_p... | 19A016_62375_4950_20165625103 | 760 | 363 | 307 | 53 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.56 | 18.690001 | 13.465702 | 15.427143 | 3.669243 | 13.463346 | 0.272488 | 4.679999 | -1.150223 | 4.115820 | 2.874086 | 2.160001 | 2.112858 | 13.465702 | 1.978568 | -0.449101 | 0.256733 | 0.146934 | -0.226983 | 0.129757 | 1.9854 | 5.620000 | 8.698000 | 10.770000 | 11.5500 | 12.328 | 13.068 | 14.370000 | 15.170000 | 15.870000 | 16.230000 | 16.573999 | 17.230000 | 17.859001 | 18.345200 | 0.695021 | 13.955336 | 14.305179 | 152 | 11238 | 621.096419 | 152.000000 | 631.573195 | 3.988847e+05 | 1.016868 | 433.00 | 13.161927 | 220.723870 | 275.177045 | 621.096419 | 193.865973 | 51.591859 | 38.322707 | 0.312135 | 0.266121 | 0.197676 | 170.240005 | 216.399994 | 238.200012 | 297.000000 | 351.00 | 387.000000 | 458.799988 | 560.0 | 649.000000 | 739.000000 | 784.00 | 844.200073 | 1001.000000 | 1144.000000 | 1447.020386 | 62.020202 | 47.763158 | 73.333333 | 307 | 363 | 40.808081 | 28.080808 | 26.842105 | 18.289474 | 41.212121 | 28.080808 | 202 | 139 | 204 | 139 | 495 | 760 | 34.615203 | |
| 9 | 2016 | 5625 | 203 | 0 | 1 | 496548 | 6239185 | 10 | Blek | 1 | 1 | 65817.664140 | 25885.144090 | 29740.796440 | 121443.60470 | 121.443605 | 5315 | 20190406 | 3000 | 160 | 11 | 1.23 | 8238 | Leica | ALS80-HP | 35 | 278000 | 42 | 100 | 62375_4950_25 | 19A016 | 43561 | 6237500 | 495000 | 19A016_62375_4950_25.laz.laz | 0 | 0 | 2 | 4147 | 815 | D:\Projekt\ForestChangeHMB\LASdata\Skanning2_p... | 19A016_62375_4950_20165625203 | 811 | 419 | 324 | 88 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.90 | 19.049999 | 12.821241 | 13.061111 | 3.807565 | 14.497555 | 0.296973 | 5.125000 | -0.701668 | 2.988488 | 3.014020 | 2.590000 | 2.618890 | 12.821241 | 2.127366 | -0.311288 | 0.239160 | 0.165925 | -0.146326 | 0.112421 | 2.7504 | 4.852000 | 7.394000 | 9.742000 | 10.6000 | 11.474 | 12.584 | 13.250000 | 14.190000 | 15.102000 | 15.725000 | 16.326000 | 17.346001 | 18.070999 | 18.723801 | 0.636807 | 13.373376 | 13.795088 | 152 | 11238 | 744.489260 | 152.000000 | 1199.026151 | 1.437664e+06 | 1.610535 | 515.00 | 7.884790 | 68.718406 | 438.186021 | 744.489260 | 312.123511 | 154.177013 | 122.548544 | 0.419245 | 0.493962 | 0.392628 | 181.080002 | 216.000000 | 245.399994 | 301.799988 | 333.00 | 361.600006 | 441.000000 | 540.0 | 661.000000 | 787.000000 | 848.00 | 921.000000 | 1188.800049 | 1383.800049 | 9459.528320 | 62.307692 | 51.664612 | 80.576923 | 324 | 419 | 41.538462 | 39.230769 | 29.099877 | 27.620222 | 45.384615 | 43.076923 | 216 | 204 | 236 | 224 | 520 | 811 | 35.084541 |
[146]:
len(predictors)
[146]:
217
Plotting the target variable
[36]:
bins = np.linspace(min(predictors['TotalAGB']),max(predictors['TotalAGB']),100)
plt.hist((predictors['TotalAGB']),bins,alpha=0.8);
Clean the dataset by removing ‘0’ AGB values
[3]:
predictors_sel = predictors.loc[(predictors['TotalAGB'] != 0)]
Plotting response variables distribution after cleaning
[4]:
bins = np.linspace(min(predictors_sel['TotalAGB']),max(predictors_sel['TotalAGB']),100)
plt.hist((predictors_sel['TotalAGB']),bins,alpha=0.8);
[106]:
#select the:
#target variable AGB
#predictor variables <- latitude, longitude, height percentiles and intensities
[5]:
AGB = predictors_sel['TotalAGB'].to_numpy()
cols = [5,6,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125]
data = predictors_sel[predictors_sel.columns[cols]]
print(data.head())
Ostkoordin Nordkoordi Elev P01 Elev P05 Elev P10 Elev P20 Elev P25 \
0 494081 6237269 1.5600 2.0295 2.379 3.278000 3.7500
1 476547 6239273 1.6846 2.1900 3.252 4.762000 5.4800
2 476282 6238964 6.9882 9.6300 11.464 13.197999 13.8750
3 476268 6239247 1.5728 1.6960 1.790 1.998000 2.1900
4 486266 6239102 2.0740 6.1450 11.005 16.719999 17.8225
Elev P30 Elev P40 Elev P50 Elev P60 Elev P70 Elev P75 p80 \
0 4.224 6.342 8.065001 10.006000 11.785000 12.265000 12.850000
1 6.000 7.080 8.740000 11.148000 15.642000 16.680000 17.459999
2 14.269 15.334 16.240000 17.120001 17.896999 18.299999 18.858000
3 2.310 2.524 2.810000 3.004000 3.220000 3.300000 3.412000
4 18.510 20.270 21.430000 22.690001 23.889999 24.289999 24.680000
Elev P90 Elev P95 Elev P99 crr Int P01 Int P05 \
0 14.574000 15.250000 16.839298 0.424836 147.119995 168.650009
1 18.514000 19.020000 19.832399 0.462006 126.000000 153.000000
2 20.077000 21.019001 22.095800 0.670032 150.000000 181.000000
3 3.900000 4.128000 4.260400 0.455759 259.320007 342.200012
4 25.360001 26.160000 26.907499 0.699449 142.649994 152.000000
Int P10 Int P20 Int P25 Int P30 Int P40 Int P50 Int P60 \
0 185.400009 207.0 216.00 229.000000 261.000000 305.5 351.000000
1 171.000000 221.0 241.00 271.000000 321.000000 389.0 461.000000
2 211.599991 289.0 343.00 399.200012 527.799927 661.5 837.400024
3 404.799988 519.0 623.00 679.000000 758.599976 834.0 921.199890
4 169.000000 189.0 199.25 207.000000 225.000000 252.0 271.000000
Int P70 Int P75 Int P80 Int P90 Int P95 Int P99
0 414.000000 450.00 495.000000 686.000061 958.000000 1302.879883
1 535.600098 588.00 658.599915 880.400146 1140.800171 1513.998901
2 1013.000000 1092.75 1235.000000 1456.000244 1666.950195 11150.000000
3 1048.200073 1196.00 1388.599854 1802.600220 2008.000000 2207.799805
4 292.000000 306.00 333.000000 519.500000 793.000000 1155.549927
[ ]:
#Explore the raw data - predictor variables
[6]:
n_plots_x = int(np.ceil(np.sqrt(data.shape[1])))+1
n_plots_y = int(np.floor(np.sqrt(data.shape[1])))
fig, ax = plt.subplots(n_plots_x, n_plots_y, figsize=(15,15), dpi=100, facecolor='w', edgecolor='k')
ax=ax.ravel()
for idx in range(data.shape[1]-1):
ax[idx].hist(data.iloc[:, idx].to_numpy().flatten())
ax[idx].set_title(data.columns[idx])
fig.tight_layout()
[107]:
#Normalize the target variable 'AGB' to range between 0 and 1 and to follow a normal distribution
[7]:
from sklearn.preprocessing import QuantileTransformer
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
qt = QuantileTransformer(
n_quantiles=500, output_distribution="normal", random_state=0
)
AGB = qt.fit_transform(AGB.reshape(-1,1))
scaler_tree = MinMaxScaler()
AGB = scaler_tree.fit_transform(AGB.reshape(-1,1)).squeeze()
plt.hist(AGB,50)
/usr/lib/python3/dist-packages/sklearn/preprocessing/_data.py:2354: UserWarning: n_quantiles (500) is greater than the total number of samples (207). n_quantiles is set to n_samples.
warnings.warn("n_quantiles (%s) is greater than the total number "
[7]:
(array([ 1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 1.,
1., 1., 3., 3., 5., 7., 9., 11., 13., 15., 17., 17., 16.,
17., 15., 13., 11., 9., 7., 5., 3., 3., 1., 1., 1., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 1.]),
array([0. , 0.02, 0.04, 0.06, 0.08, 0.1 , 0.12, 0.14, 0.16, 0.18, 0.2 ,
0.22, 0.24, 0.26, 0.28, 0.3 , 0.32, 0.34, 0.36, 0.38, 0.4 , 0.42,
0.44, 0.46, 0.48, 0.5 , 0.52, 0.54, 0.56, 0.58, 0.6 , 0.62, 0.64,
0.66, 0.68, 0.7 , 0.72, 0.74, 0.76, 0.78, 0.8 , 0.82, 0.84, 0.86,
0.88, 0.9 , 0.92, 0.94, 0.96, 0.98, 1. ]),
<a list of 50 Patch objects>)
[108]:
#Normalize the predictor variables to range between 0 and 1
[8]:
#Normalize the data
scaler_data = MinMaxScaler()
data_transformed = scaler_data.fit_transform(data)
n_plots_x = int(np.ceil(np.sqrt(data.shape[1])))+1
n_plots_y = int(np.floor(np.sqrt(data.shape[1])))
fig, ax = plt.subplots(n_plots_x, n_plots_y, figsize=(10, 10), dpi=100, facecolor='w', edgecolor='k')
ax=ax.ravel()
for idx in range(data.shape[1]-1):
ax[idx].hist(data_transformed[:,idx].flatten())
ax[idx].set_title(data.columns[idx])
fig.tight_layout()
[62]:
#Extract the column headers for later plotting the feature importance
featx = (data.columns.values)
2.10.3. Random Forest Regression (RFR)
[9]:
# Data splitting into train and test subsets
X_train_rf, X_test_rf, Y_train_rf, Y_test_rf = train_test_split(data_transformed,AGB, test_size=0.25, random_state=24)
y_train_rf = np.ravel(Y_train_rf)
y_test_rf = np.ravel(Y_test_rf)
[10]:
# Define the RFR model
rfReg = RandomForestRegressor(min_samples_leaf=15, oob_score=True)
rfReg.fit(X_train_rf, y_train_rf)
dic_pred_RF = {}
dic_pred_RF['train'] = rfReg.predict(X_train_rf)
dic_pred_RF['test'] = rfReg.predict(X_test_rf)
pearsonr_train_RF = np.corrcoef(dic_pred_RF['train'],y_train_rf)**2
pearsonr_test_RF = np.corrcoef(dic_pred_RF['test'],y_test_rf)**2
pearsonr_all_RF = [pearsonr_train_RF[0,1],pearsonr_test_RF[0,1]]
pearsonr_all_RF
[10]:
[0.5850426153769053, 0.634618398346184]
[11]:
# checking the oob score
rfReg.oob_score_
[11]:
0.4477686796959137
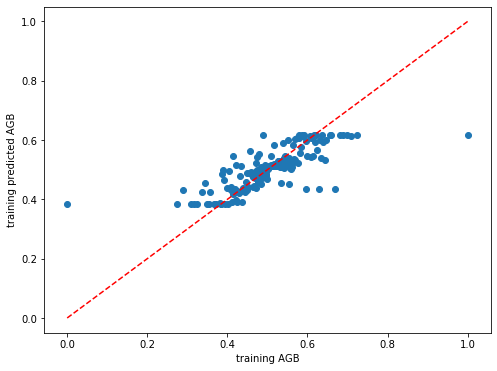
[481]:
# Plot the model performance for the training set
plt.rcParams["figure.figsize"] = (8,6)
plt.scatter(y_train_rf,dic_pred_RF['train'])
plt.xlabel('training AGB')
plt.ylabel('training predicted AGB')
ident = [0, 1]
plt.plot(ident,ident,'r--')
[481]:
[<matplotlib.lines.Line2D at 0x7f47ae1d04c0>]
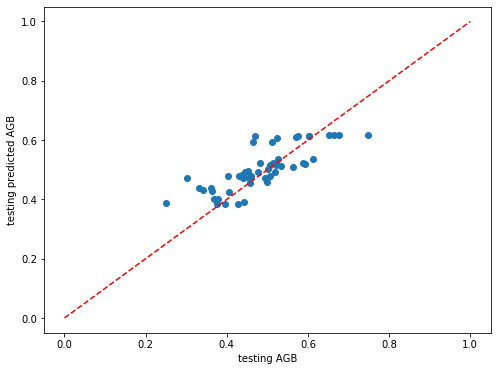
[482]:
# Plot the model performance for the test set
plt.rcParams["figure.figsize"] = (8,6)
plt.scatter(y_test_rf,dic_pred_RF['test'])
plt.xlabel('testing AGB')
plt.ylabel('testing predicted AGB')
ident = [0, 1]
plt.plot(ident,ident,'r--')
[482]:
[<matplotlib.lines.Line2D at 0x7f47ae409520>]
[ ]:
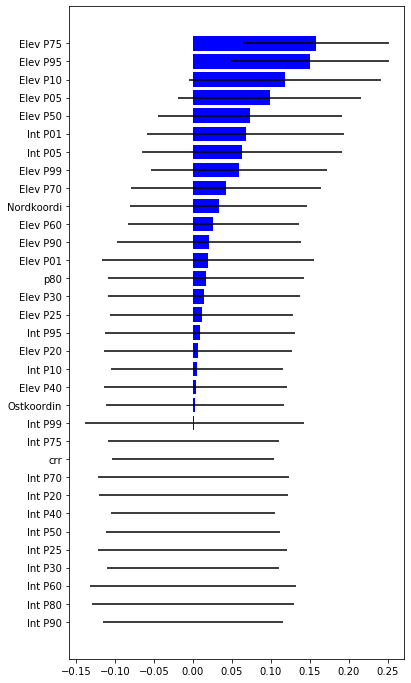
# Compute the feature importance for the predictor variables
[12]:
impt = [rfReg.feature_importances_, np.std([tree.feature_importances_ for tree in rfReg.estimators_],axis=1)]
ind = np.argsort(impt[0])
[13]:
ind
[13]:
array([30, 29, 26, 23, 22, 25, 24, 21, 27, 17, 28, 32, 0, 8, 20, 5, 31,
6, 7, 13, 2, 14, 10, 1, 11, 16, 19, 18, 9, 3, 4, 15, 12])
[64]:
plt.rcParams["figure.figsize"] = (6,12)
plt.barh(range(len(featx)),impt[0][ind],color="b", xerr=impt[1][ind], align="center")
plt.yticks(range(len(featx)),featx[ind]);
[65]:
# Model statistics
print('Mean Absolute Error (MAE):', metrics.mean_absolute_error(Y_test_rf, dic_pred_RF['test']))
print('Mean Squared Error (MSE):', metrics.mean_squared_error(Y_test_rf, dic_pred_RF['test']))
print('Root Mean Squared Error (RMSE):', metrics.mean_squared_error(Y_test_rf, dic_pred_RF['test'], squared=False))
print('R^2:', metrics.r2_score(Y_test_rf, dic_pred_RF['test']))
Mean Absolute Error (MAE): 0.04844966911077575
Mean Squared Error (MSE): 0.003917523367916487
Root Mean Squared Error (RMSE): 0.06259012196757957
R^2: 0.5984579817055615
2.10.3.1. Support Vector Machine for Regression (SVR)
[115]:
# Dataset splitting into train and test data subsets
X_train_svr, X_test_svr, y_train_svr, y_test_svr = train_test_split(data_transformed,AGB, test_size=0.50, random_state=24)
print('X_train.shape:{}, X_test.shape:{} '.format(X_train_svr.shape, X_test_svr.shape))
scaler = MinMaxScaler()
X_train_svr = scaler.fit_transform(X_train_svr)
X_test_svr = scaler.transform(X_test_svr)
X_train.shape:(103, 33), X_test.shape:(104, 33)
[116]:
# Define the SVR model
svr = SVR(kernel='linear')
svr.fit(X_train_svr, y_train_svr) # Fit the SVR model according to the given training data.
print('Accuracy of SVR on training set: {:.5f}'.format(svr.score(X_train_svr, y_train_svr))) # Returns the coefficient of determination (R^2) of the prediction.
print('Accuracy of SVR on test set: {:.5f}'.format(svr.score(X_test_svr, y_test_svr)))
Accuracy of SVR on training set: 0.62139
Accuracy of SVR on test set: 0.56197
[117]:
# Predict for the train and the test datasets
dic_pred_svr = {}
dic_pred_svr['train'] = svr.predict(X_train_svr)
dic_pred_svr['test'] = svr.predict(X_test_svr)
[118]:
# Model statistics
print('Root Mean Squared Error (RMSE):', metrics.mean_squared_error(dic_pred_svr['test'], y_test_svr, squared=False))
print('R^2:', metrics.r2_score(y_test_svr,dic_pred_svr['test']))
Root Mean Squared Error (RMSE): 0.07149401467860782
R^2: 0.5619712935228112
[185]:
#on increasing/ decreasing the test_size the svr.score was increasing but the train score was going down
2.10.4. Feedfoward neural network (FF-NN)
[71]:
# Set a random seed
seed=29
torch.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
np.random.seed(seed)
[72]:
#Split the dataset into train and test subsets
X_train, X_test, y_train, y_test = train_test_split(data_transformed,AGB, test_size=0.7, random_state=0)
X_train = torch.FloatTensor(X_train)
y_train = torch.FloatTensor(y_train)
X_test = torch.FloatTensor(X_test)
y_test = torch.FloatTensor(y_test)
print('X_train.shape: {}, X_test.shape: {}, y_train.shape: {}, y_test.shape: {}'.format(X_train.shape, X_test.shape, y_train.shape, y_test.shape))
print('X_train.min: {}, X_test.shape: {}, y_train.shape: {}, y_test.shape: {}'.format(X_train.min(), X_test.min(), y_train.min(), y_test.min()))
print('X_train.max: {}, X_test.shape: {}, y_train.shape: {}, y_test.shape: {}'.format(X_train.max(), X_test.max(), y_train.max(), y_test.max()))
X_train.shape: torch.Size([62, 33]), X_test.shape: torch.Size([145, 33]), y_train.shape: torch.Size([62]), y_test.shape: torch.Size([145])
X_train.min: 0.0, X_test.shape: 0.0, y_train.shape: 0.0, y_test.shape: 0.25131121277809143
X_train.max: 1.0, X_test.shape: 1.0, y_train.shape: 0.7247803211212158, y_test.shape: 1.0
[73]:
# Define the FeedForward Neural Network
class Feedforward(torch.nn.Module):
def __init__(self, input_size, hidden_size, output_size=1):
super(Feedforward, self).__init__()
self.input_size = input_size
self.hidden_size = hidden_size
self.fc1 = torch.nn.Linear(self.input_size, self.hidden_size)
self.fc2 = torch.nn.Linear(self.hidden_size, self.hidden_size)
# self.fc3 = torch.nn.Linear(self.hidden_size, self.hidden_size)
# self.fc4 = torch.nn.Linear(self.hidden_size, self.hidden_size)
self.relu = torch.nn.ReLU()
self.fc3 = torch.nn.Linear(self.hidden_size, output_size)
self.sigmoid = torch.nn.Sigmoid()
#self.tanh = torch.nn.Tanh()
def forward(self, x):
hidden = self.relu(self.fc1(x))
hidden = self.relu(self.fc2(hidden))
# hidden = self.relu(self.fc3(hidden))
# hidden = self.relu(self.fc4(hidden))
output = self.sigmoid(self.fc3(hidden))
return output
[74]:
# FF-NN model.train()
epochs = 4000
hid_dim_range = [512]#,256
lr_range = [0.01]#,0.5,1]
#Let's create a place to save these models, so we can
path_to_save_models = './models'
if not os.path.exists(path_to_save_models):
os.makedirs(path_to_save_models)
for hid_dim in hid_dim_range:
for lr in lr_range:
print('\nhid_dim: {}, lr: {}'.format(hid_dim, lr))
if 'model' in globals():
print('Deleting previous model')
del model, criterion, optimizer
model = Feedforward(33, hid_dim)
criterion = torch.nn.MSELoss()
optimizer = torch.optim.SGD(model.parameters(), lr = lr)
all_loss_train=[]
all_loss_val=[]
for epoch in range(epochs):
model.train()
optimizer.zero_grad()
# Forward pass
y_pred = model(X_train)
# Compute Loss
loss = criterion(y_pred.squeeze(), y_train)
# Backward pass
loss.backward()
optimizer.step()
all_loss_train.append(loss.item())
model.eval()
with torch.no_grad():
y_pred = model(X_test)
# Compute Loss
loss = criterion(y_pred.squeeze(), y_test)
all_loss_val.append(loss.item())
if epoch%100==0:
y_pred = y_pred.detach().numpy().squeeze()
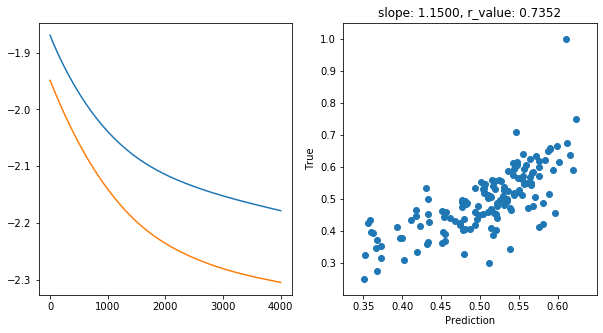
slope, intercept, r_value, p_value, std_err = scipy.stats.linregress(y_pred, y_test)
print('Epoch {}, train_loss: {:.4f}, val_loss: {:.4f}, r_value: {:.4f}'.format(epoch,all_loss_train[-1],all_loss_val[-1],r_value))
fig,ax=plt.subplots(1,2,figsize=(10,5))
ax[0].plot(np.log10(all_loss_train))
ax[0].plot(np.log10(all_loss_val))
ax[1].scatter(y_pred, y_test)
ax[1].set_xlabel('Prediction')
ax[1].set_ylabel('True')
ax[1].set_title('slope: {:.4f}, r_value: {:.4f}'.format(slope, r_value))
plt.show()
name_to_save = os.path.join(path_to_save_models,'model_SGD_' + str(epochs) + '_lr' + str(lr) + '_hid_dim' + str(hid_dim))
print('Saving model to ', name_to_save)
model_state = {
'epoch': epoch + 1,
'state_dict': model.state_dict(),
'optimizer' : optimizer.state_dict(),
}
torch.save(model_state, name_to_save +'.pt')
hid_dim: 512, lr: 0.01
Epoch 0, train_loss: 0.0135, val_loss: 0.0112, r_value: -0.5676
Epoch 100, train_loss: 0.0128, val_loss: 0.0106, r_value: 0.1132
Epoch 200, train_loss: 0.0121, val_loss: 0.0101, r_value: 0.5879
Epoch 300, train_loss: 0.0116, val_loss: 0.0096, r_value: 0.6581
Epoch 400, train_loss: 0.0111, val_loss: 0.0091, r_value: 0.6764
Epoch 500, train_loss: 0.0107, val_loss: 0.0087, r_value: 0.6844
Epoch 600, train_loss: 0.0103, val_loss: 0.0084, r_value: 0.6887
Epoch 700, train_loss: 0.0100, val_loss: 0.0080, r_value: 0.6915
Epoch 800, train_loss: 0.0097, val_loss: 0.0077, r_value: 0.6936
Epoch 900, train_loss: 0.0094, val_loss: 0.0075, r_value: 0.6956
Epoch 1000, train_loss: 0.0091, val_loss: 0.0072, r_value: 0.6972
Epoch 1100, train_loss: 0.0089, val_loss: 0.0070, r_value: 0.6988
Epoch 1200, train_loss: 0.0087, val_loss: 0.0068, r_value: 0.7003
Epoch 1300, train_loss: 0.0085, val_loss: 0.0066, r_value: 0.7017
Epoch 1400, train_loss: 0.0084, val_loss: 0.0065, r_value: 0.7031
Epoch 1500, train_loss: 0.0082, val_loss: 0.0063, r_value: 0.7045
Epoch 1600, train_loss: 0.0081, val_loss: 0.0062, r_value: 0.7058
Epoch 1700, train_loss: 0.0080, val_loss: 0.0061, r_value: 0.7071
Epoch 1800, train_loss: 0.0079, val_loss: 0.0060, r_value: 0.7084
Epoch 1900, train_loss: 0.0078, val_loss: 0.0059, r_value: 0.7096
Epoch 2000, train_loss: 0.0077, val_loss: 0.0058, r_value: 0.7109
Epoch 2100, train_loss: 0.0076, val_loss: 0.0057, r_value: 0.7121
Epoch 2200, train_loss: 0.0075, val_loss: 0.0057, r_value: 0.7135
Epoch 2300, train_loss: 0.0074, val_loss: 0.0056, r_value: 0.7148
Epoch 2400, train_loss: 0.0074, val_loss: 0.0055, r_value: 0.7162
Epoch 2500, train_loss: 0.0073, val_loss: 0.0055, r_value: 0.7175
Epoch 2600, train_loss: 0.0073, val_loss: 0.0054, r_value: 0.7189
Epoch 2700, train_loss: 0.0072, val_loss: 0.0054, r_value: 0.7203
Epoch 2800, train_loss: 0.0071, val_loss: 0.0053, r_value: 0.7217
Epoch 2900, train_loss: 0.0071, val_loss: 0.0053, r_value: 0.7231
Epoch 3000, train_loss: 0.0070, val_loss: 0.0052, r_value: 0.7244
Epoch 3100, train_loss: 0.0070, val_loss: 0.0052, r_value: 0.7257
Epoch 3200, train_loss: 0.0069, val_loss: 0.0052, r_value: 0.7270
Epoch 3300, train_loss: 0.0069, val_loss: 0.0051, r_value: 0.7282
Epoch 3400, train_loss: 0.0069, val_loss: 0.0051, r_value: 0.7294
Epoch 3500, train_loss: 0.0068, val_loss: 0.0051, r_value: 0.7306
Epoch 3600, train_loss: 0.0068, val_loss: 0.0051, r_value: 0.7318
Epoch 3700, train_loss: 0.0067, val_loss: 0.0050, r_value: 0.7329
Epoch 3800, train_loss: 0.0067, val_loss: 0.0050, r_value: 0.7341
Epoch 3900, train_loss: 0.0067, val_loss: 0.0050, r_value: 0.7352
Saving model to ./models/model_SGD_4000_lr0.01_hid_dim512
[75]:
# Define the function to compute model statistics
def make_preds(X_train,y_train,X_test,y_test):
trainPredict = model(X_train)
testPredict = model(X_test)
trainScore = np.sqrt(mean_squared_error(y_train, trainPredict))
print('Train Score: %.2f RMSE' % (trainScore))
#print('Train R^2: ', r2_score(y_train, trainPredict))
testScore = np.sqrt(mean_squared_error(y_test, testPredict))
#print('Train R^2: ', r2_score(y_test, testPredict))
print('TMetrics RMSE: %.2f RMSE' % (testScore))
return trainPredict, testPredict
[76]:
# Model statistics
model.eval()
with torch.no_grad():
make_preds(X_train,y_train.reshape(-1,1),X_test,y_test.reshape(-1,1))
Train Score: 0.08 RMSE
TMetrics RMSE: 0.07 RMSE
[77]:
# To compute the feature importance for the predictor variables
import shap
test_set= X_test
test_labels= y_test
explainer = shap.DeepExplainer(model, X_test)
shap_values = explainer.shap_values(test_set)
Using a non-full backward hook when the forward contains multiple autograd Nodes is deprecated and will be removed in future versions. This hook will be missing some grad_input. Please use register_full_backward_hook to get the documented behavior.
[100]:
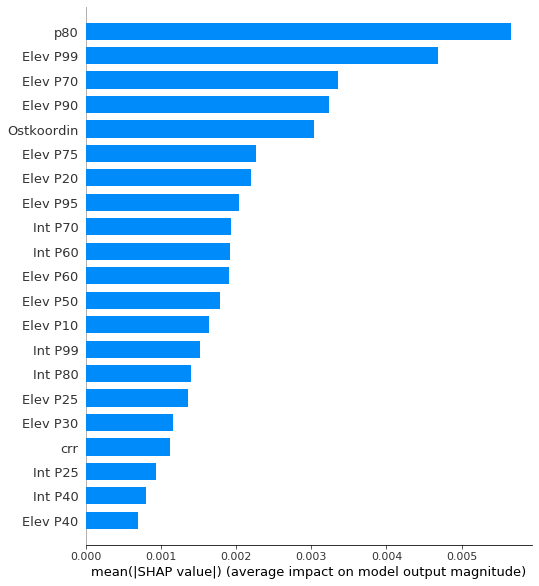
#SHAP global interpretation
# The summary plot shows the most important features and the magnitude of their impact on the model. It is the global interpretation.
print(shap_values.shape)
shap.summary_plot(shap_values, plot_type = 'bar', feature_names = data.columns, max_display=21)
(145, 33)
[119]:
# Tabular rerpesentation of the model statistics summary of the above 3 models used (RFR, SVR, FF-NN)
print(tabulate([['RFR', round(metrics.mean_squared_error(y_test_rf, dic_pred_RF['test'], squared=False),2),
round(metrics.r2_score(y_test_rf, dic_pred_RF['test']),2)], ['SVR', round(metrics.mean_squared_error(y_test_svr, dic_pred_svr['test'], squared=False),2),
round(metrics.r2_score(y_test_svr,dic_pred_svr['test']),2)], ['FeedF NN', '0.07', 0.59]], headers=['Algorithm', 'RMSE', 'R^2'])) #['GLM', 63.49, 0.45]
Algorithm RMSE R^2
----------- ------ -----
RFR 0.06 0.6
SVR 0.07 0.56
FeedF NN 0.07 0.59
[120]:
# RFR and FF-NN was performing very similar with R^2 and RMSE values either similar/higher/lower compared to eachother.
#Therefore, the data represented in the above table is for one random instance.
2.10.4.1. Predict the raster using RFR
[81]:
# Define RFR with the 2 predictor variables corresponding to the available rasters
X_rf2 = predictors_sel.iloc[:,[86,90]].values
Y_rf2 = predictors_sel.iloc[:,15:16].values
X_train_rf2, X_test_rf2, Y_train_rf2, Y_test_rf2 = train_test_split(X_rf2, Y_rf2, test_size=0.25, random_state=24)
y_train_rf2 = np.ravel(Y_train_rf2)
y_test_rf2 = np.ravel(Y_test_rf2)
rfReg_rf2 = RandomForestRegressor(min_samples_leaf=20, oob_score=True)
rfReg_rf2.fit(X_train_rf2, y_train_rf2);
dic_pred_rf2 = {}
dic_pred_rf2['train'] = rfReg_rf2.predict(X_train_rf2)
dic_pred_rf2['test'] = rfReg_rf2.predict(X_test_rf2)
#print('Mean Absolute Error (MAE):', metrics.mean_absolute_error(y_test_rf2, dic_pred_rf2['test']))
#print('Mean Squared Error (MSE):', metrics.mean_squared_error(y_test_rf2, dic_pred_rf2['test']))
#print('Root Mean Squared Error (RMSE):', metrics.mean_squared_error(y_test_rf2, dic_pred_rf2['test'], squared=False))
#print('R^2:', metrics.r2_score(y_test_rf2, dic_pred_rf2['test']))
Mean Absolute Error (MAE): 41.108253168570315
Mean Squared Error (MSE): 2699.6832371086425
Root Mean Squared Error (RMSE): 51.95847608531877
R^2: 0.6374537638503947
[82]:
# Bash commands to crop the rasters for extracting a sub-area from the study region
#%%bash
#gdalwarp -cutline sub-area.shp -crop_to_cutline -dstalpha P80_t2.tif p80_t2_crop.tif
#gdalwarp -cutline sub-area.shp -crop_to_cutline -dstalpha CanopyReliefRatio_t2.tif crr_t2_crop.tif
[83]:
# Import rasters
p80 = rasterio.open("../images/p80_t2_crop.tif")
crr = rasterio.open("../images/crr_t2_crop.tif")
[84]:
# Stack the 2 raster layers correspong to the predictor variables
#predictors_rasters = [p80,crr]
stack = Raster(p80,crr)
[85]:
# Check the dimension of the raster stack to be given as input to the RFR
stack.count
[85]:
2
[86]:
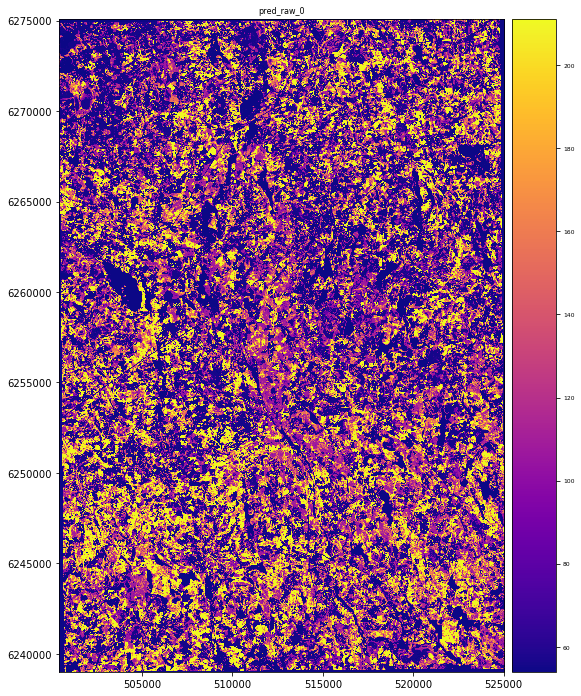
# Predict the raster for the target variable 'AGB'
result = stack.predict(estimator=rfReg_rf2, dtype='int16', nodata=-1)
print(result)
<pyspatialml.raster.Raster object at 0x7f7588ea6820>
[87]:
# Plot the resultin g map
plt.rcParams["figure.figsize"] = (12,12)
result.iloc[0].cmap = "plasma"
result.plot()
plt.show()
[ ]: